7RXS
 
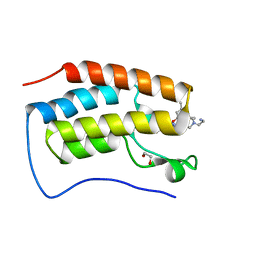 | | Crystal of BRD4(D1) with 2-[(3S)-3-{5-[2-(3,5-dimethylphenoxy)pyrimidin-4-yl]-4-(4-iodophenyl)-1H-imidazol-1-yl}pyrrolidin-1-yl]ethan-1-amine | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3S)-3-{5-[2-(3,5-dimethylphenoxy)pyrimidin-4-yl]-4-(4-iodophenyl)-1H-imidazol-1-yl}pyrrolidin-1-yl]ethan-1-amine, Bromodomain-containing protein 4 | | Authors: | Cui, H, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-23 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | A Structure-based Design Approach for Generating High Affinity BRD4 D1-Selective Chemical Probes.
J.Med.Chem., 65, 2022
|
|
7RXT
 
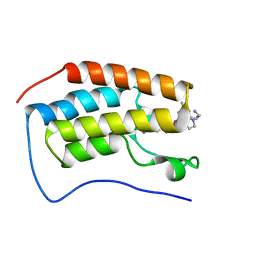 | | Crystal of BRD4(D1) with 2-[(3R)-3-{5-[2-(3,5-dimethylphenoxy)pyrimidin-4-yl]-4-(4-iodophenyl)-1H-imidazol-1-yl}pyrrolidin-1-yl]ethan-1-amine | | Descriptor: | 2-[(3R)-3-{5-[2-(3,5-dimethylphenoxy)pyrimidin-4-yl]-4-(4-iodophenyl)-1H-imidazol-1-yl}pyrrolidin-1-yl]ethan-1-amine, Bromodomain-containing protein 4 | | Authors: | Cui, H, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-23 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | A Structure-based Design Approach for Generating High Affinity BRD4 D1-Selective Chemical Probes.
J.Med.Chem., 65, 2022
|
|
7RXR
 
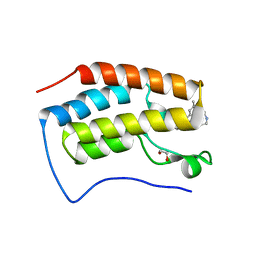 | | Crystal Structure of BRD4(D1) with 4-[4-(4-bromophenyl)-1-(piperidin-4-yl)-1H-imidazol-5-yl]-N-(3,5-dimethylphenyl)pyrimidin-2-amine | | Descriptor: | 1,2-ETHANEDIOL, 4-[4-(4-bromophenyl)-1-(piperidin-4-yl)-1H-imidazol-5-yl]-N-(3,5-dimethylphenyl)pyrimidin-2-amine, Bromodomain-containing protein 4 | | Authors: | Cui, H, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-23 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | A Structure-based Design Approach for Generating High Affinity BRD4 D1-Selective Chemical Probes.
J.Med.Chem., 65, 2022
|
|
6XF5
 
 | | Cryo-EM structure of a biotinylated SARS-CoV-2 spike probe in the prefusion state (RBDs down) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Cerutti, G, Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-02 | | Last modified: | 2020-12-02 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure-Based Design with Tag-Based Purification and In-Process Biotinylation Enable Streamlined Development of SARS-CoV-2 Spike Molecular Probes.
SSRN, 2020
|
|
7SFP
 
 | |
7SFN
 
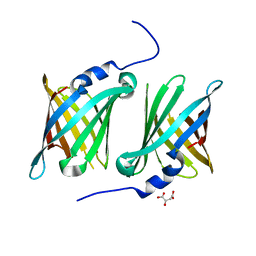 | |
6Y4Q
 
 | | Structure of a stapled peptide bound to MDM2 | | Descriptor: | ACE-LEU-THR-PHE-GLY-GLU-TYR-TRP-ALA-GLN-LEU-ALA-SER, E3 ubiquitin-protein ligase Mdm2, ~{N}-[(1-ethyl-1,2,3-triazol-4-yl)methyl]-~{N},5-dimethyl-4-[2-[2-methyl-5-[methyl-[(1-propyl-1,2,3-triazol-4-yl)methyl]carbamoyl]thiophen-3-yl]cyclopenten-1-yl]thiophene-2-carboxamide | | Authors: | Pantelejevs, T, Bakanovych, I. | | Deposit date: | 2020-02-22 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Diarylethene moiety as an enthalpy-entropy switch: photoisomerizable stapled peptides for modulating p53/MDM2 interaction.
Org.Biomol.Chem., 18, 2020
|
|
7RYT
 
 | |
6ASP
 
 | | Structure of Grp94 with methyl 3-chloro-2-(2-(1-(2-ethoxybenzyl)-1 H-imidazol-2-yl)ethyl)-4,6-dihydroxybenzoate, a Grp94-selective inhibitor and promising therapeutic lead for treating myocilin-associated glaucoma | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Endoplasmin, GLYCEROL, ... | | Authors: | Huard, D.J.E, Lieberman, R.L. | | Deposit date: | 2017-08-25 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.696 Å) | | Cite: | Trifunctional High-Throughput Screen Identifies Promising Scaffold To Inhibit Grp94 and Treat Myocilin-Associated Glaucoma.
ACS Chem. Biol., 13, 2018
|
|
6AO9
 
 | |
6AOA
 
 | |
6AOB
 
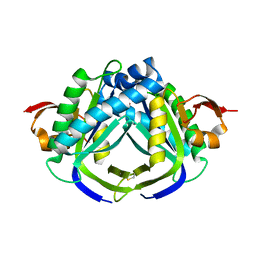 | |
6ASQ
 
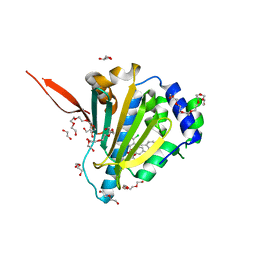 | | Structure of Grp94 bound to methyl 2-[2-(2-benzylpyridin-3-yl)ethyl]-3-chloro-4,6-dihydroxybenzoate, a pan-Hsp90 inhibitor | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, DI(HYDROXYETHYL)ETHER, Endoplasmin, ... | | Authors: | Huard, D.J.E, Lieberman, R.L. | | Deposit date: | 2017-08-25 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Trifunctional High-Throughput Screen Identifies Promising Scaffold To Inhibit Grp94 and Treat Myocilin-Associated Glaucoma.
ACS Chem. Biol., 13, 2018
|
|
7TUM
 
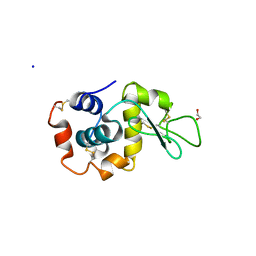 | | Multi-Hit SFX using MHz XFEL sources- first hit | | Descriptor: | 1,2-ETHANEDIOL, Lysozyme C, SODIUM ION | | Authors: | Darmanin, C, Holmes, S, Abbey, B. | | Deposit date: | 2022-02-03 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Megahertz pulse trains enable multi-hit serial femtosecond crystallography experiments at X-ray free electron lasers.
Nat Commun, 13, 2022
|
|
6CPD
 
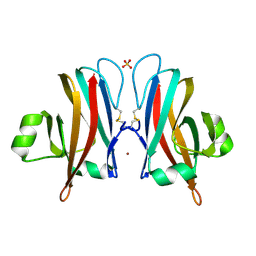 | |
6CXH
 
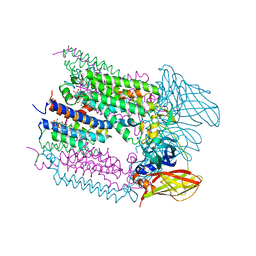 | |
6KYF
 
 | | Crystal structure of an anti-CRISPR protein | | Descriptor: | AcrF11, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Niu, Y, Wang, H, Zhang, Y, Feng, Y. | | Deposit date: | 2019-09-18 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | A Type I-F Anti-CRISPR Protein Inhibits the CRISPR-Cas Surveillance Complex by ADP-Ribosylation.
Mol.Cell, 80, 2020
|
|
6MH4
 
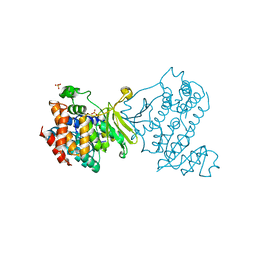 | |
6MJV
 
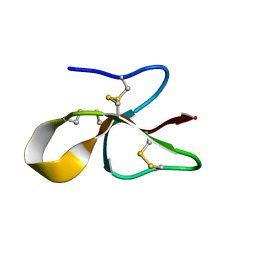 | | A consensus human beta defensin | | Descriptor: | Human beta-defensin | | Authors: | Amero, C, Villegas-Moreno, J, Rodriguez, A, Norton, R.S, Corzo, G. | | Deposit date: | 2018-09-22 | | Release date: | 2019-08-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Antimicrobial activity and structure of a consensus human beta-defensin and its comparison to a novel putative hBD10.
Proteins, 88, 2020
|
|
4AGO
 
 | | Structure of the p53 core domain mutant Y220C bound to the stabilizing small molecule PhiKan5174 | | Descriptor: | CELLULAR TUMOR ANTIGEN P53, TERT-BUTYL [3-(3-{[4-(DIETHYLAMINO)PIPERIDIN-1-YL]METHYL}-4-HYDROXY-5-IODOPHENYL)PROP-2-YN-1-YL]CARBAMATE, ZINC ION | | Authors: | Joerger, A.C, Wilcken, R, Fersht, A.R, Boeckler, F.M. | | Deposit date: | 2012-01-30 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Halogen-Enriched Fragment Libraries as Leads for Drug Rescue of Mutant P53.
J.Am.Chem.Soc., 134, 2012
|
|
4AGN
 
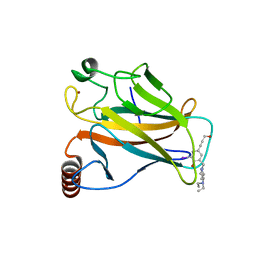 | | Structure of the p53 core domain mutant Y220C bound to the stabilizing small molecule PhiKan5116 | | Descriptor: | 2-{[4-(DIETHYLAMINO)PIPERIDIN-1-YL]METHYL}-4-(3-HYDROXYPROP-1-YN-1-YL)-6-IODOPHENOL, CELLULAR TUMOR ANTIGEN P53, ZINC ION | | Authors: | Joerger, A.C, Wilcken, R, Boeckler, F.M, Fersht, A.R. | | Deposit date: | 2012-01-30 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Halogen-Enriched Fragment Libraries as Leads for Drug Rescue of Mutant P53.
J.Am.Chem.Soc., 134, 2012
|
|
6MW5
 
 | | UDP-galactose:glucoside-Skp1 alpha-D-galactosyltransferase with bound UDP and Platinum | | Descriptor: | 1,2-ETHANEDIOL, PLATINUM (II) ION, UDP-galactose:glucoside-Skp1 alpha-D-galactosyltransferase, ... | | Authors: | Kim, H.W, Wood, Z.A, West, C.M. | | Deposit date: | 2018-10-29 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A terminal alpha 3-galactose modification regulates an E3 ubiquitin ligase subunit in Toxoplasma gondii .
J.Biol.Chem., 295, 2020
|
|
6MH5
 
 | |
4AGQ
 
 | | Structure of the p53 core domain mutant Y220C bound to the stabilizing small molecule PhiKan5196 | | Descriptor: | 2-{[4-(diethylamino)piperidin-1-yl]methyl}-6-iodo-4-[3-(phenylamino)prop-1-yn-1-yl]phenol, CELLULAR TUMOR ANTIGEN P53, ZINC ION | | Authors: | Joerger, A.C, Wilcken, R, Boeckler, F.M, Fersht, A.R. | | Deposit date: | 2012-01-30 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Halogen-Enriched Fragment Libraries as Leads for Drug Rescue of Mutant P53.
J.Am.Chem.Soc., 134, 2012
|
|
6MW8
 
 | | UDP-galactose:glucoside-Skp1 alpha-D-galactosyltransferase with bound UDP and Manganese | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, MANGANESE (II) ION, ... | | Authors: | Kim, H.W, Wood, Z.A, West, C.M. | | Deposit date: | 2018-10-29 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.756 Å) | | Cite: | A terminal alpha 3-galactose modification regulates an E3 ubiquitin ligase subunit in Toxoplasma gondii .
J.Biol.Chem., 295, 2020
|
|
