1GJY
 
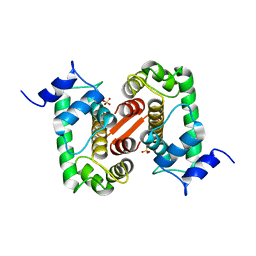 | | The X-ray structure of the Sorcin Calcium Binding Domain (SCBD) provides insight into the phosphorylation and calcium dependent processess | | Descriptor: | SORCIN, SULFATE ION | | Authors: | Ilari, A, Johnson, K.A, Nastopoulos, V, Tsernoglou, D, Chiancone, E. | | Deposit date: | 2001-08-06 | | Release date: | 2002-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of the Sorcin Calcium Binding Domain Provides a Model of Ca(2+)-Dependent Processes in the Full-Length Protein
J.Mol.Biol., 317, 2002
|
|
1KFC
 
 | | CRYSTAL STRUCTURE OF ALPHAT183V MUTANT OF TRYPTOPHAN SYNTHASE FROM SALMONELLA TYPHIMURIUM With Indole Propanol Phosphate | | Descriptor: | INDOLE-3-PROPANOL PHOSPHATE, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Kulik, V, Weyand, M, Siedel, R, Niks, D, Arac, D, Dunn, M.F, Schlichting, I. | | Deposit date: | 2001-11-20 | | Release date: | 2003-01-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | On the Role of AlphaTHR183 in the Allosteric Regulation and Catalytic Mechanism of Tryptophan Synthase
J.Mol.Biol., 324, 2002
|
|
1KFE
 
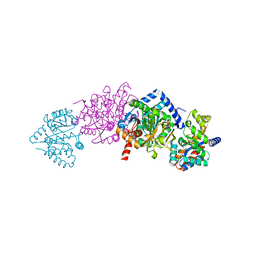 | | CRYSTAL STRUCTURE OF ALPHAT183V MUTANT OF TRYPTOPHAN SYNTHASE FROM SALMONELLA TYPHIMURIUM WITH L-Ser Bound To The Beta Site | | Descriptor: | SODIUM ION, TRYPTOPHAN SYNTHASE ALPHA CHAIN, TRYPTOPHAN SYNTHASE BETA CHAIN, ... | | Authors: | Kulik, V, Weyand, M, Siedel, R, Niks, D, Arac, D, Dunn, M.F, Schlichting, I. | | Deposit date: | 2001-11-20 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | On the Role of AlphaTHR183 in the Allosteric Regulation and Catalytic Mechanism of Tryptophan Synthase
J.Mol.Biol., 324, 2002
|
|
1SS6
 
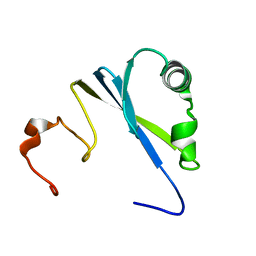 | | Solution structure of SEP domain from human p47 | | Descriptor: | NSFL1 cofactor p47 | | Authors: | Soukenik, M, Leidert, M, Sievert, V, Buessow, K, Leitner, D, Labudde, D, Ball, L.J, Oschkinat, H. | | Deposit date: | 2004-03-23 | | Release date: | 2004-11-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The SEP domain of p47 acts as a reversible competitive inhibitor of cathepsin L
FEBS Lett., 576, 2004
|
|
1KV6
 
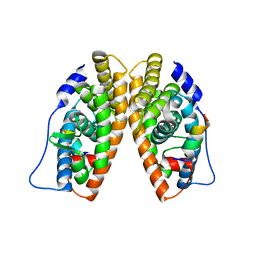 | | X-ray structure of the orphan nuclear receptor ERR3 ligand-binding domain in the constitutively active conformation | | Descriptor: | ESTROGEN-RELATED RECEPTOR GAMMA, steroid receptor coactivator 1 | | Authors: | Greschik, H, Wurtz, J.-M, Sanglier, S, Bourguet, W, van Dorsselaer, A, Moras, D, Renaud, J.-P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2002-01-25 | | Release date: | 2003-01-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Functional Evidence for Ligand-Independent Transcriptional Activation by the Estrogen-Related Receptor 3
Mol.Cell, 9, 2002
|
|
1KJ2
 
 | | Murine Alloreactive ScFv TCR-Peptide-MHC Class I Molecule Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Allogeneic H-2Kb MHC Class I Molecule, Beta-2 microglobulin, ... | | Authors: | Reiser, J.-B, Gregoire, C, Darnault, C, Mosser, T, Guimezanes, A, Schmitt-Verhulst, A.-M, Fontecilla-Camps, J.C, Mazza, G, Malissen, B, Housset, D. | | Deposit date: | 2001-12-04 | | Release date: | 2002-03-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | A T cell receptor CDR3beta loop undergoes conformational changes of unprecedented magnitude upon binding to a peptide/MHC class I complex.
Immunity, 16, 2002
|
|
1KN7
 
 | | Solution structure of the tandem inactivation domain (residues 1-75) of potassium channel RCK4 (Kv1.4) | | Descriptor: | VOLTAGE-GATED POTASSIUM CHANNEL PROTEIN KV1.4 | | Authors: | Wissmann, R, Bildl, W, Oliver, D, Beyermann, M, Kalbitzer, H.R, Bentrop, D, Fakler, B. | | Deposit date: | 2001-12-18 | | Release date: | 2003-05-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Function of the "Tandem Inactivation Domain" of the Neuronal A-type
Potassium Channel Kv1.4
J.Biol.Chem., 278, 2003
|
|
1KFJ
 
 | | CRYSTAL STRUCTURE OF WILD-TYPE TRYPTOPHAN SYNTHASE COMPLEXED WITH L-SERINE | | Descriptor: | SODIUM ION, TRYPTOPHAN SYNTHASE ALPHA CHAIN, TRYPTOPHAN SYNTHASE BETA CHAIN, ... | | Authors: | Kulik, V, Weyand, M, Seidel, R, Niks, D, Arac, D, Dunn, M.F, Schlichting, I. | | Deposit date: | 2001-11-21 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | On the role of alphaThr183 in the allosteric regulation and catalytic mechanism of tryptophan synthase.
J.Mol.Biol., 324, 2002
|
|
1KFB
 
 | | CRYSTAL STRUCTURE OF ALPHAT183V MUTANT OF TRYPTOPHAN SYNTHASE FROM SALMONELLA TYPHIMURIUM WITH Indole Glycerol Phosphate | | Descriptor: | INDOLE-3-GLYCEROL PHOSPHATE, PYRIDOXAL-5'-PHOSPHATE, TRYPTOPHAN SYNTHASE ALPHA CHAIN, ... | | Authors: | Kulik, V, Weyand, M, Siedel, R, Niks, D, Arac, D, Dunn, M.F, Schlichting, I. | | Deposit date: | 2001-11-20 | | Release date: | 2003-01-07 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | On the Role of alphaTHR183 in the Allosteric Regulation and Catalytic Mechanism of Tryptophan Synthase
J.Mol.Biol., 324, 2002
|
|
1KFK
 
 | | Crystal structure of Tryptophan Synthase From Salmonella Typhimurium | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, TRYPTOPHAN SYNTHASE ALPHA CHAIN, ... | | Authors: | Kulik, V, Weyand, M, Seidel, R, Niks, D, Arac, D, Dunn, M.F, Schlichting, I. | | Deposit date: | 2001-11-21 | | Release date: | 2003-10-21 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | On the role of alphaThr183 in the allosteric regulation and catalytic mechanism of tryptophan synthase.
J.Mol.Biol., 324, 2002
|
|
1KJ3
 
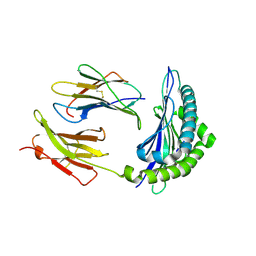 | | Mhc Class I H-2Kb molecule complexed with pKB1 peptide | | Descriptor: | BETA-2 MICROGLOBULIN, H-2KB MHC CLASS I MOLECULE ALPHA CHAIN, NATURALLY PROCESSED OCTAPEPTIDE PKB1 | | Authors: | Reiser, J.-B, Gregoire, C, Darnault, C, Mosser, T, Guimezanes, A, Schmitt-Verhulst, A.-M, Fontecilla-Camps, J.C, Mazza, G, Malissen, B, Housset, D. | | Deposit date: | 2001-12-04 | | Release date: | 2002-03-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A T cell receptor CDR3beta loop undergoes conformational changes of unprecedented magnitude upon binding to a peptide/MHC class I complex.
Immunity, 16, 2002
|
|
1G2G
 
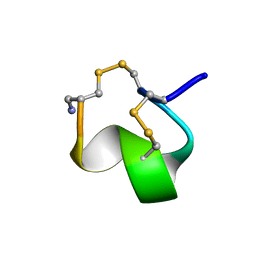 | | MINIMAL CONFORMATION OF THE ALPHA-CONOTOXIN IMI FOR THE ALPHA7 NEURONAL NICOTINIC ACETYLCHOLINE RECEPTOR RECOGNITION | | Descriptor: | ALPHA-CONOTOXIN IMI | | Authors: | Lamthanh, H, Jegou-Matheron, C, Servent, D, Menez, A, Lancelin, J.M. | | Deposit date: | 2000-10-19 | | Release date: | 2000-11-08 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Minimal conformation of the alpha-conotoxin ImI for the alpha7 neuronal nicotinic acetylcholine receptor recognition: correlated CD, NMR and binding studies.
FEBS Lett., 454, 1999
|
|
1KHM
 
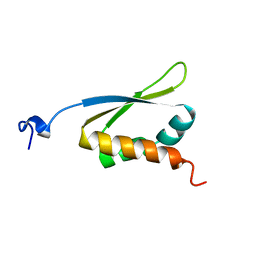 | | C-TERMINAL KH DOMAIN OF HNRNP K (KH3) | | Descriptor: | PROTEIN (HNRNP K) | | Authors: | Baber, J, Libutti, D, Levens, D, Tjandra, N. | | Deposit date: | 1999-01-07 | | Release date: | 2000-01-12 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | High precision solution structure of the C-terminal KH domain of heterogeneous nuclear ribonucleoprotein K, a c-myc transcription factor.
J.Mol.Biol., 289, 1999
|
|
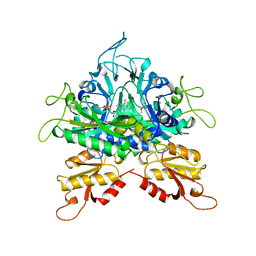
1GGM
 
 | |
7RXL
 
 | |
2P64
 
 | | D domain of b-TrCP | | Descriptor: | CADMIUM ION, F-box/WD repeat protein 1A | | Authors: | Neculai, D, Orlicky, S, Ceccarelli, D. | | Deposit date: | 2007-03-16 | | Release date: | 2007-06-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Suprafacial orientation of the SCFCdc4 dimer accommodates multiple geometries for substrate ubiquitination.
Cell(Cambridge,Mass.), 129, 2007
|
|
8RLZ
 
 | |
6F18
 
 | | Structure of Mb NMH H64V, V68A mutant complex with EDA | | Descriptor: | ETHYL ACETATE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tinzl, M, Hayashi, T, Mori, T, Hilvert, D. | | Deposit date: | 2017-11-21 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Capture and characterization of a reactive haem-carbenoid complex in an artificial metalloenzyme
Nat Catal, 1, 2018
|
|
2AAT
 
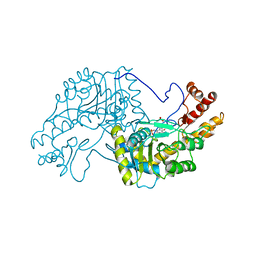 | | 2.8-ANGSTROMS-RESOLUTION CRYSTAL STRUCTURE OF AN ACTIVE-SITE MUTANT OF ASPARTATE AMINOTRANSFERASE FROM ESCHERICHIA COLI | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ASPARTATE AMINOTRANSFERASE, SULFATE ION | | Authors: | Smith, D, Almo, S.C, Toney, M, Ringe, D. | | Deposit date: | 1989-05-30 | | Release date: | 1989-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 2.8-A-resolution crystal structure of an active-site mutant of aspartate aminotransferase from Escherichia coli.
Biochemistry, 28, 1989
|
|
8RQL
 
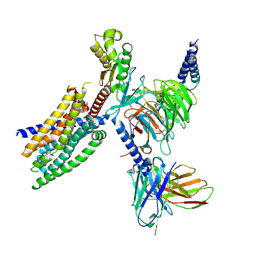 | | TAS2R14 receptor bound to flufenamic acid and gustducin | | Descriptor: | 2-[[3-(TRIFLUOROMETHYL)PHENYL]AMINO] BENZOIC ACID, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Matzov, D, Peri, L, Niv, M, Shalev Benami, M. | | Deposit date: | 2024-01-18 | | Release date: | 2024-11-27 | | Last modified: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | A bitter anti-inflammatory drug binds at two distinct sites of a human bitter taste GPCR.
Nat Commun, 15, 2024
|
|
7PB8
 
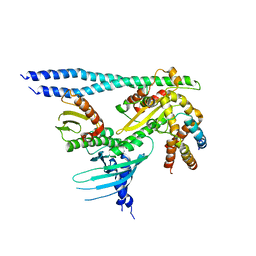 | | Crystal structure of the CENP-OPQUR complex | | Descriptor: | Centromere protein O, Centromere protein P, Centromere protein Q, ... | | Authors: | Bellini, D, Yatskevich, S, Muir, K.W, Barford, D. | | Deposit date: | 2021-07-31 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.68 Å) | | Cite: | Structure of the human inner kinetochore bound to a centromeric CENP-A nucleosome.
Science, 376, 2022
|
|
7PB4
 
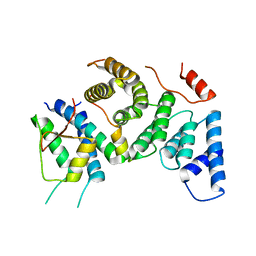 | | Cenp-HIK 3-protein complex | | Descriptor: | Centromere protein H, Centromere protein I, Centromere protein K | | Authors: | Bellini, D, Yatskevich, S, Muir, W.K, Barford, D. | | Deposit date: | 2021-07-30 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure of the human inner kinetochore bound to a centromeric CENP-A nucleosome.
Science, 376, 2022
|
|
7PKN
 
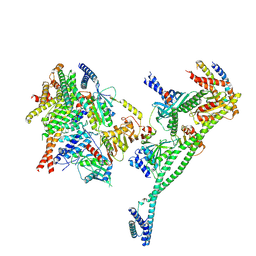 | | Structure of the human CCAN deltaCT complex | | Descriptor: | Centromere protein H, Centromere protein I, Centromere protein K, ... | | Authors: | Muir, K.W, Yatskevich, S, Bellini, D, Barford, D. | | Deposit date: | 2021-08-25 | | Release date: | 2022-04-27 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the human inner kinetochore bound to a centromeric CENP-A nucleosome.
Science, 376, 2022
|
|
6UC5
 
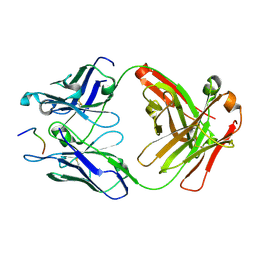 | | Fab397 in complex with NPNA peptide | | Descriptor: | Fab397 heavy chain, Fab397 light chain, NPNA peptide | | Authors: | Pholcharee, T, Oyen, D, Wilson, I.A. | | Deposit date: | 2019-09-13 | | Release date: | 2020-01-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Diverse Antibody Responses to Conserved Structural Motifs in Plasmodium falciparum Circumsporozoite Protein.
J.Mol.Biol., 432, 2020
|
|
6FKC
 
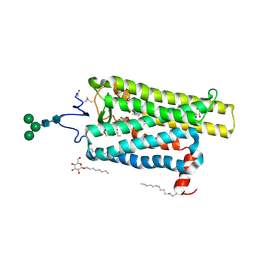 | | Crystal structure of N2C/D282C stabilized opsin bound to RS15 | | Descriptor: | 3-[1'-[(2~{S})-2-(4-chlorophenyl)-3-methyl-butanoyl]spiro[1,3-benzodioxole-2,4'-piperidine]-5-yl]propanoic acid, PALMITIC ACID, Rhodopsin, ... | | Authors: | Mattle, D, Standfuss, J, Dawson, R. | | Deposit date: | 2018-01-23 | | Release date: | 2018-04-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Ligand channel in pharmacologically stabilized rhodopsin.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
