4CJN
 
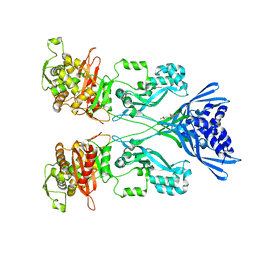 | | Crystal structure of PBP2a from MRSA in complex with quinazolinone ligand | | Descriptor: | (E)-3-(2-(4-cyanostyryl)-4-oxoquinazolin-3(4H)-yl)benzoic acid, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Bouley, R, Otero, L.H, Rojas-Altuve, A, Hermoso, J.A. | | Deposit date: | 2013-12-21 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.947 Å) | | Cite: | Discovery of Antibiotic (E)-3-(3-Carboxyphenyl)-2-(4-Cyanostyryl)Quinazolin-4(3H)-One.
J.Am.Chem.Soc., 137, 2015
|
|
5Y2T
 
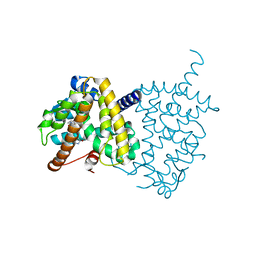 | | Structure of PPARgamma ligand binding domain - lobeglitazone complex | | Descriptor: | (5S)-5-[[4-[2-[[6-(4-methoxyphenoxy)pyrimidin-4-yl]-methyl-amino]ethoxy]phenyl]methyl]-1,3-thiazolidine-2,4-dione, Peroxisome proliferator-activated receptor gamma | | Authors: | Im, Y.J, Lee, M. | | Deposit date: | 2017-07-27 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of PPAR gamma complexed with lobeglitazone and pioglitazone reveal key determinants for the recognition of antidiabetic drugs
Sci Rep, 7, 2017
|
|
5Y2O
 
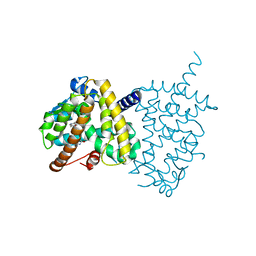 | | Structure of PPARgamma ligand binding domain-pioglitazone complex | | Descriptor: | (5S)-5-[[4-[2-(5-ethylpyridin-2-yl)ethoxy]phenyl]methyl]-1,3-thiazolidine-2,4-dione, Peroxisome proliferator-activated receptor gamma | | Authors: | Im, Y.J, Lee, M. | | Deposit date: | 2017-07-26 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structures of PPAR gamma complexed with lobeglitazone and pioglitazone reveal key determinants for the recognition of antidiabetic drugs
Sci Rep, 7, 2017
|
|
6I0A
 
 | |
6I09
 
 | |
6I0N
 
 | |
6I05
 
 | |
1E9Z
 
 | |
1E9Y
 
 | | Crystal structure of Helicobacter pylori urease in complex with acetohydroxamic acid | | Descriptor: | ACETOHYDROXAMIC ACID, NICKEL (II) ION, UREASE SUBUNIT ALPHA, ... | | Authors: | Ha, N.-C, Oh, S.-T, Oh, B.-H. | | Deposit date: | 2000-11-01 | | Release date: | 2001-11-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Supramolecular Assembly and Acid Resistance of Helicobacter Pylori Urease
Nat.Struct.Biol., 8, 2001
|
|
6C8C
 
 | | Chimeric Pol kappa RIR Rev1 C-terminal domain in complex with JHRE06 | | Descriptor: | 8-chloro-2-[(2,4-dichlorophenyl)amino]-3-(3-methylbutanoyl)-5-nitroquinolin-4(1H)-one, Chimeric protein of the Pol Kappa RIR helix and the Rev1 C-terminal domain | | Authors: | Najeeb, J, Zhou, P. | | Deposit date: | 2018-01-24 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Small Molecule Targeting Mutagenic Translesion Synthesis Improves Chemotherapy.
Cell, 178, 2019
|
|
6C59
 
 | | Chimeric Pol kappa RIR Rev1 C-terminal domain | | Descriptor: | ACETATE ION, Chimeric Pol kappa RIR Rev1 C-terminal domain | | Authors: | Wojtaszek, J.L, Zhou, P. | | Deposit date: | 2018-01-15 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A Small Molecule Targeting Mutagenic Translesion Synthesis Improves Chemotherapy.
Cell, 178, 2019
|
|
5AA2
 
 | | Crystal structure of MltF from Pseudomonas aeruginosa in complex with NAM-pentapeptide. | | Descriptor: | CHLORIDE ION, MEMBRANE-BOUND LYTIC MUREIN TRANSGLYCOSYLASE F, N-ACETYLGLUCOSAMINE-1,6-ANHYDRO-N-ACETYLMURAMIC ACID L-ALA-D-GLU-M-DAP-D-ALA-D-ALA | | Authors: | Dominguez-Gil, T, Acebron, I, Hermoso, J.A. | | Deposit date: | 2015-07-23 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Activation by Allostery in Cell-Wall Remodeling by a Modular Membrane-Bound Lytic Transglycosylase from Pseudomonas aeruginosa.
Structure, 24, 2016
|
|
5AA1
 
 | | Crystal structure of MltF from Pseudomonas aeruginosa in complex with NAG-anhNAM-pentapeptide | | Descriptor: | CHLORIDE ION, MEMBRANE-BOUND LYTIC MUREIN TRANSGLYCOSYLASE F, N-ACETYLGLUCOSAMINE-1,6-ANHYDRO-N-ACETYLMURAMIC ACID L-ALA-D-GLU-M-DAP-D-ALA-D-ALA | | Authors: | Dominguez-Gil, T, Acebron, I, Hermoso, J.A. | | Deposit date: | 2015-07-23 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Activation by Allostery in Cell-Wall Remodeling by a Modular Membrane-Bound Lytic Transglycosylase from Pseudomonas aeruginosa.
Structure, 24, 2016
|
|
5CA5
 
 | | Structure of the C. elegans NONO-1 homodimer | | Descriptor: | 1,2-ETHANEDIOL, NONO-1 | | Authors: | Knott, G.J, Bond, C.S. | | Deposit date: | 2015-06-29 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Caenorhabditis elegans NONO-1: Insights into DBHS protein structure, architecture, and function.
Protein Sci., 24, 2015
|
|
5AA4
 
 | | Crystal structure of MltF from Pseudomonas aeruginosa in complex with cell-wall tetrapeptide | | Descriptor: | MEMBRANE-BOUND LYTIC MUREIN TRANSGLYCOSYLASE F, [6-[[(2~{R})-1-azanyl-1-oxidanylidene-propan-2-yl]amino]-6-oxidanylidene-5-[[(4~{R})-5-oxidanyl-5-oxidanylidene-4-[[(2~{S})-2-[[(2~{R})-2-oxidanylpropanoyl]amino]propanoyl]amino]pentanoyl]amino]hexyl]azanium | | Authors: | Dominguez-Gil, T, Acebron, I, Hermoso, J.A. | | Deposit date: | 2015-07-23 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Activation by Allostery in Cell-Wall Remodeling by a Modular Membrane-Bound Lytic Transglycosylase from Pseudomonas aeruginosa.
Structure, 24, 2016
|
|
5A5X
 
 | |
5CTV
 
 | | Catalytic domain of LytA, the major autolysin of Streptococcus pneumoniae, (C60A, H133A, C136A mutant) complexed with peptidoglycan fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-beta-D-glucopyranoside-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-beta-D-glucopyranoside, Autolysin, fragment of peptidoglycan | | Authors: | Achour, A, Sandalova, T, Mellroth, P. | | Deposit date: | 2015-07-24 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | The crystal structure of the major pneumococcal autolysin LytA in complex with a large peptidoglycan fragment reveals the pivotal role of glycans for lytic activity.
Mol.Microbiol., 101, 2016
|
|
4OZS
 
 | | RNA binding protein | | Descriptor: | Alpha solenoid protein | | Authors: | Gully, B.S, Bond, C.S. | | Deposit date: | 2014-02-19 | | Release date: | 2015-04-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | The design and structural characterization of a synthetic pentatricopeptide repeat protein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3E1F
 
 | |
3DYM
 
 | | E. coli (lacZ) beta-galactosidase (H418E) | | Descriptor: | Beta-galactosidase, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Juers, D.H, Huber, R.E, Matthews, B.W. | | Deposit date: | 2008-07-28 | | Release date: | 2008-10-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Direct and indirect roles of His-418 in metal binding and in the activity of beta-galactosidase (E. coli).
Protein Sci., 18, 2009
|
|
3DYP
 
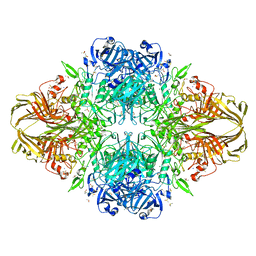 | | E. coli (lacZ) beta-galactosidase (H418N) | | Descriptor: | Beta-galactosidase, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Juers, D.H, Huber, R.E, Matthews, B.W. | | Deposit date: | 2008-07-28 | | Release date: | 2008-10-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Direct and indirect roles of His-418 in metal binding and in the activity of beta-galactosidase (E. coli).
Protein Sci., 18, 2009
|
|
3DYO
 
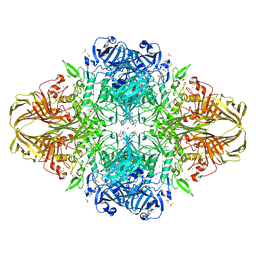 | | E. coli (lacZ) beta-galactosidase (H418N) in complex with IPTG | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Juers, D.H, Huber, R.E, Matthews, B.W. | | Deposit date: | 2008-07-28 | | Release date: | 2008-10-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct and indirect roles of His-418 in metal binding and in the activity of beta-galactosidase (E. coli).
Protein Sci., 18, 2009
|
|
3KM5
 
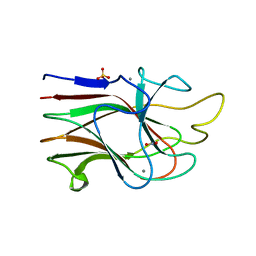 | | Crystal Structure Analysis of the K2 Cleaved Adhesin Domain of Lys-gingipain (Kgp) | | Descriptor: | CALCIUM ION, GLYCEROL, Lysine specific cysteine protease, ... | | Authors: | Li, N, Collyer, C.A, Hunter, N. | | Deposit date: | 2009-11-09 | | Release date: | 2010-03-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure determination and analysis of a haemolytic gingipain adhesin domain from Porphyromonas gingivalis
Mol.Microbiol., 76, 2010
|
|
4IWT
 
 | |
4IVV
 
 | |
