7XTU
 
 | | The structure of TfCut S130A | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Yang, Y, Jiang, P.C, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Complete bio-degradation of poly(butylene adipate-co-terephthalate) via engineered cutinases.
Nat Commun, 14, 2023
|
|
7XTS
 
 | | The apo structure of the engineered TfCut S130A | | Descriptor: | SODIUM ION, alpha/beta hydrolase | | Authors: | Yang, Y, Jiang, P.C, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Complete bio-degradation of poly(butylene adipate-co-terephthalate) via engineered cutinases.
Nat Commun, 14, 2023
|
|
7XTW
 
 | | The structure of IsPETase in complex with MHET | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-hydroxyethyloxycarbonyl)benzoic acid, GLYCEROL, ... | | Authors: | Yang, Y, Jiang, P.C, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Complete bio-degradation of poly(butylene adipate-co-terephthalate) via engineered cutinases.
Nat Commun, 14, 2023
|
|
7XTR
 
 | | The apo structure of the engineered TfCut | | Descriptor: | GLYCEROL, LITHIUM ION, SULFATE ION, ... | | Authors: | Yang, Y, Jiang, P.C, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Complete bio-degradation of poly(butylene adipate-co-terephthalate) via engineered cutinases.
Nat Commun, 14, 2023
|
|
7XTT
 
 | | The structure of engineered TfCut S130A in complex with MHET | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-hydroxyethyloxycarbonyl)benzoic acid, SODIUM ION, ... | | Authors: | Yang, Y, Jiang, P.C, Huang, J.W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Complete bio-degradation of poly(butylene adipate-co-terephthalate) via engineered cutinases.
Nat Commun, 14, 2023
|
|
2HWG
 
 | | Structure of phosphorylated Enzyme I of the phosphoenolpyruvate:sugar phosphotransferase system | | Descriptor: | MAGNESIUM ION, OXALATE ION, Phosphoenolpyruvate-protein phosphotransferase | | Authors: | Lim, K, Teplyakov, A, Herzberg, O. | | Deposit date: | 2006-08-01 | | Release date: | 2006-11-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of phosphorylated enzyme I, the phosphoenolpyruvate:sugar phosphotransferase system sugar translocation signal protein.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
5H9D
 
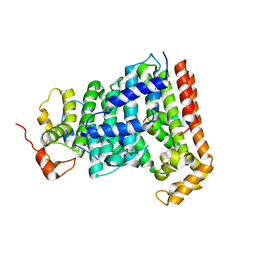 | | Crystal structure of Heptaprenyl Diphosphate Synthase from Staphylococcus aureus | | Descriptor: | C-terminal peptide from Heptaprenyl diphosphate synthase (HEPPP synthase) subunit 1 family protein, Farnesyl pyrophosphate synthetase, Heptaprenyl diphosphate synthase (HEPPP synthase) subunit 1 family protein, ... | | Authors: | Wei, H.L, Liu, W.D, Zheng, Y.Y, Ko, T.P, Chen, C.C, Guo, R.T. | | Deposit date: | 2015-12-28 | | Release date: | 2016-12-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structure, Function, and Inhibition of Staphylococcus aureus Heptaprenyl Diphosphate Synthase
ChemMedChem, 11, 2016
|
|
8IBI
 
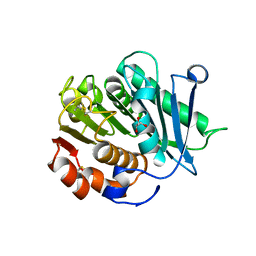 | | Inactive mutant of CtPL-H210S/F214I | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Li, X, Shi, B.L, Zeng, Z.Y, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-02-10 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Functional tailoring of a PET hydrolytic enzyme expressed in Pichia pastoris.
Bioresour Bioprocess, 10, 2023
|
|
8IBJ
 
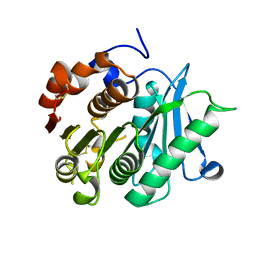 | | Inactive mutant of CtPL-H210S/F214I/N181A/F235L | | Descriptor: | PET hydrolase | | Authors: | Li, X, Shi, B.L, Zeng, Z.Y, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-02-10 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Functional tailoring of a PET hydrolytic enzyme expressed in Pichia pastoris.
Bioresour Bioprocess, 10, 2023
|
|
8IAN
 
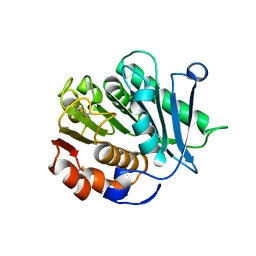 | | Crystal structure of CtPL-H210S/F214I mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, PET hydrolase | | Authors: | Li, X, Shi, B.L, Zeng, Z.Y, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-02-08 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Functional tailoring of a PET hydrolytic enzyme expressed in Pichia pastoris.
Bioresour Bioprocess, 10, 2023
|
|
8IHS
 
 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 in complex with ochratoxin A | | Descriptor: | (2~{S})-2-[[(3~{R})-5-chloranyl-3-methyl-8-oxidanyl-1-oxidanylidene-3,4-dihydroisochromen-7-yl]carbonylamino]-3-phenyl-propanoic acid, Amidohydrolase family protein, ZINC ION | | Authors: | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-02-23 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
8IHR
 
 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 in complex with Phe | | Descriptor: | Amidohydrolase family protein, PHENYLALANINE, ZINC ION | | Authors: | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-02-23 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
8IHQ
 
 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 | | Descriptor: | Amidohydrolase family protein, ZINC ION | | Authors: | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-02-23 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
8J85
 
 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 mutant S88E in complex with ochratoxin A | | Descriptor: | (2~{S})-2-[[(3~{R})-5-chloranyl-3-methyl-8-oxidanyl-1-oxidanylidene-3,4-dihydroisochromen-7-yl]carbonylamino]-3-phenyl-propanoic acid, Amidohydrolase family protein, ZINC ION | | Authors: | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-04-30 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
6JCN
 
 | | Yeast dehydrodolichyl diphosphate synthase complex subunit NUS1 | | Descriptor: | Dehydrodolichyl diphosphate synthase complex subunit NUS1, SULFATE ION | | Authors: | Ko, T.-P, Ma, J, Liu, W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2019-01-29 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural insights to heterodimeric cis-prenyltransferases through yeast dehydrodolichyl diphosphate synthase subunit Nus1.
Biochem.Biophys.Res.Commun., 515, 2019
|
|
6KV9
 
 | | MoeE5 in complex with UDP-glucuronic acid and NAD | | Descriptor: | MoeE5, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, URIDINE-5'-DIPHOSPHATE-GLUCURONIC ACID | | Authors: | Ko, T.-P, Liu, W, Sun, H, Liu, W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2019-09-03 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structure of an antibiotic-synthesizing UDP-glucuronate 4-epimerase MoeE5 in complex with substrate.
Biochem.Biophys.Res.Commun., 521, 2020
|
|
6KVC
 
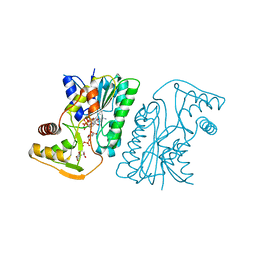 | | MoeE5 in complex with UDP-glucose and NAD | | Descriptor: | MoeE5, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Ko, T.-P, Liu, W, Sun, H, Liu, W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2019-09-04 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure of an antibiotic-synthesizing UDP-glucuronate 4-epimerase MoeE5 in complex with substrate.
Biochem.Biophys.Res.Commun., 521, 2020
|
|
5CJB
 
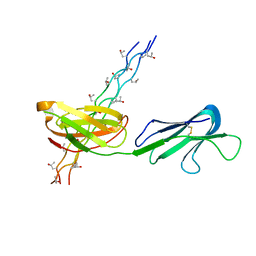 | | Human Osteoclast Associated Receptor (OSCAR) in complex with a collagen-like peptide | | Descriptor: | Osteoclast-associated immunoglobulin-like receptor, collagen-like peptide | | Authors: | Gao, G.F, Qi, J, Shi, Y, Haywood, J. | | Deposit date: | 2015-07-14 | | Release date: | 2016-01-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | Structural basis of collagen recognition by human osteoclast-associated receptor and design of osteoclastogenesis inhibitors.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5CJ8
 
 | |
5D4Y
 
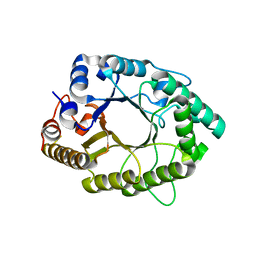 | | A psychrophilic glycoside hydrolase family 10 endo-beta-1,4-xylanase | | Descriptor: | beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, xylanase | | Authors: | Zheng, Y, Guo, R.T. | | Deposit date: | 2015-08-10 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insight into potential cold adaptation mechanism through a psychrophilic glycoside hydrolase family 10 endo-beta-1,4-xylanase.
J.Struct.Biol., 193, 2016
|
|
7W5G
 
 | | The apo structure of trichobrasilenol synthase TaTC6 with the space group of orthorhombic | | Descriptor: | GLYCEROL, SULFATE ION, Terpene cyclase 6 | | Authors: | Chen, C, Wang, T, Yang, Y, Zhang, L, Ko, T, Huang, J, Guo, R. | | Deposit date: | 2021-11-30 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the cyclization of unusual brasilane-type sesquiterpenes.
Int.J.Biol.Macromol., 209, 2022
|
|
7W5F
 
 | | The apo structure of trichobrasilenol synthase TaTC6 with the space group of monoclinic | | Descriptor: | MAGNESIUM ION, Terpene cyclase 6 | | Authors: | Chen, C, Wang, T, Yang, Y, Zhang, L, Ko, T, Huang, J, Guo, R. | | Deposit date: | 2021-11-30 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural insights into the cyclization of unusual brasilane-type sesquiterpenes.
Int.J.Biol.Macromol., 209, 2022
|
|
7W5I
 
 | | The structure of trichobrasilenol synthase TaTC6 in complex with FPP-1 | | Descriptor: | FARNESYL DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Chen, C, Wang, T, Yang, Y, Zhang, L, Ko, T, Huang, J, Guo, R. | | Deposit date: | 2021-11-30 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural insights into the cyclization of unusual brasilane-type sesquiterpenes.
Int.J.Biol.Macromol., 209, 2022
|
|
7W5J
 
 | | The structure of trichobrasilenol synthase TaTC6 in complex with FPP-2 | | Descriptor: | FARNESYL, FARNESYL DIPHOSPHATE, GLYCEROL, ... | | Authors: | Chen, C, Wang, T, Yang, Y, Zhang, L, Ko, T, Huang, J, Guo, R. | | Deposit date: | 2021-11-30 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural insights into the cyclization of unusual brasilane-type sesquiterpenes.
Int.J.Biol.Macromol., 209, 2022
|
|
7W5H
 
 | | The structure of trichobrasilenol synthase TaTC6 in complex with FsPP | | Descriptor: | GLYCEROL, MAGNESIUM ION, MALONATE ION, ... | | Authors: | Chen, C, Wang, T, Yang, Y, Zhang, L, Ko, T, Huang, J, Guo, R. | | Deposit date: | 2021-11-30 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural insights into the cyclization of unusual brasilane-type sesquiterpenes.
Int.J.Biol.Macromol., 209, 2022
|
|
