5CRM
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, DIMETHYL SULFOXIDE, ... | | Authors: | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | Deposit date: | 2015-07-23 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
5CQT
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | Descriptor: | Bromodomain-containing protein 4, N-cyclohexyl-1-ethyl-2-oxo-1,2-dihydrobenzo[cd]indole-6-sulfonamide | | Authors: | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | Deposit date: | 2015-07-22 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
5CY9
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, GLYCEROL, ... | | Authors: | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | Deposit date: | 2015-07-30 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
5CRZ
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-chloro-N-(1-ethyl-2-oxo-1,2-dihydrobenzo[cd]indol-6-yl)-4-fluorobenzenesulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | Deposit date: | 2015-07-23 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
5CTL
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | Descriptor: | Bromodomain-containing protein 4, N-(1-ethyl-2-oxo-1,2-dihydrobenzo[cd]indol-6-yl)benzenesulfonamide | | Authors: | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | Deposit date: | 2015-07-24 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
5DX4
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with benzo[cd]indol-2(1H)-one ligand | | Descriptor: | 1,2-ETHANEDIOL, 5-bromo-N-(1-ethyl-2-oxo-1,2-dihydrobenzo[cd]indol-6-yl)-2-methoxybenzenesulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Zhang, Y, Song, M, Liu, Z, Xue, X, Xu, Y. | | Deposit date: | 2015-09-23 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones as Potent and Specific BET Bromodomain Inhibitors: Structure-Based Virtual Screening, Optimization, and Biological Evaluation
J.Med.Chem., 59, 2016
|
|
6O5W
 
 | | Crystal structure of MORC3 CW domain fused with viral influenza A NS1 peptide | | Descriptor: | NS1-linked peptide,MORC family CW-type zinc finger protein 3, ZINC ION | | Authors: | Ahn, J, Zhang, Y, Vann, K.R, Kutateleadze, T. | | Deposit date: | 2019-03-04 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.412 Å) | | Cite: | MORC3 Is a Target of the Influenza A Viral Protein NS1.
Structure, 27, 2019
|
|
6P79
 
 | | Engineered single chain antibody C9+C14 ScFv | | Descriptor: | Engineered antibody heavy chain, Engineered antibody light chain | | Authors: | Zhang, Y, Li, W, Marshall, N. | | Deposit date: | 2019-06-05 | | Release date: | 2020-04-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.583 Å) | | Cite: | Computer-based Engineering of Thermostabilized Antibody Fragments.
Aiche J, 66, 2020
|
|
3PCU
 
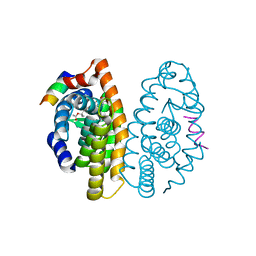 | | Crystal structure of human retinoic X receptor alpha ligand-binding domain complexed with LX0278 and SRC1 peptide | | Descriptor: | 2-[(2S)-6-(2-methylbut-3-en-2-yl)-7-oxo-2,3-dihydro-7H-furo[3,2-g]chromen-2-yl]propan-2-yl acetate, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Zhang, H, Zhang, Y, Shen, H, Chen, J, Li, C, Chen, L, Hu, L, Jiang, H, Shen, X. | | Deposit date: | 2010-10-22 | | Release date: | 2011-11-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | (+)-Rutamarin as a Dual Inducer of Both GLUT4 Translocation and Expression Efficiently Ameliorates Glucose Homeostasis in Insulin-Resistant Mice.
Plos One, 7, 2012
|
|
2QCX
 
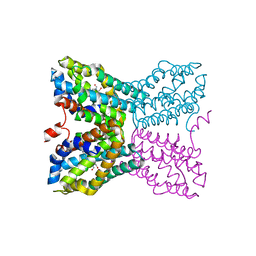 | | Crystal structure of Bacillus subtilis TenA Y112F mutant complexed with formyl aminomethyl pyrimidine | | Descriptor: | N-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-N-(2-HYDROXYETHYL)FORMAMIDE, Transcriptional activator tenA | | Authors: | Zhang, Y, Jenkins, A.L, Begley, T.P, Ealick, S.E. | | Deposit date: | 2007-06-19 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutagenesis studies on TenA: A thiamin salvage enzyme from Bacillus subtilis
Bioorg.Chem., 36, 2008
|
|
2Q5A
 
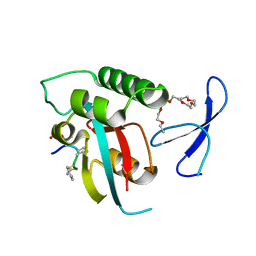 | | human Pin1 bound to L-PEPTIDE | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE, Five residue peptide, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Noel, J.P, Zhang, Y. | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for high-affinity peptide inhibition of human Pin1.
Acs Chem.Biol., 2, 2007
|
|
2VI5
 
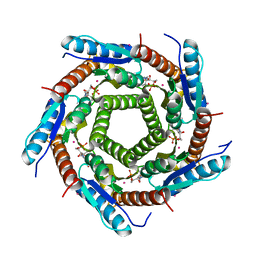 | | LUMAZINE SYNTHASE FROM MYCOBACTERIUM TUBERCULOSIS BOUND TO N-6-(ribitylamino)pyrimidine-2,4(1H,3H)-dione-5-yl-propionamide | | Descriptor: | 1-deoxy-1-{[(5S)-2,6-dioxo-5-(propanoylamino)-1,2,5,6-tetrahydropyrimidin-4-yl]amino}-D-ribitol, 6,7-DIMETHYL-8-RIBITYLLUMAZINE SYNTHASE, PHOSPHATE ION, ... | | Authors: | Morgunova, E, Zhang, Y, Jin, G, Illarionov, B, Bacher, A, Fischer, M, Cushman, M, Ladenstein, R. | | Deposit date: | 2007-11-27 | | Release date: | 2008-04-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A New Series of N-[2,4-Dioxo-6-D-Ribitylamino-1,2, 3,4-Tetrahydropyrimidin-5-Yl]Oxalamic Acid Derivatives as Inhibitors of Lumazine Syntase and Riboflavin Synthase: Design, Synthesis, Biochemical Evaluation, Crystallography and Mechanistic Implications.
J.Org.Chem., 73, 2008
|
|
5D4C
 
 | |
5D4D
 
 | |
5D4E
 
 | |
5LS6
 
 | | Structure of Human Polycomb Repressive Complex 2 (PRC2) with inhibitor | | Descriptor: | 1-[(1~{R})-1-[1-[2,2-bis(fluoranyl)propyl]piperidin-4-yl]ethyl]-~{N}-[(4-methoxy-6-methyl-2-oxidanylidene-3~{H}-pyridin-3-yl)methyl]-2-methyl-indole-3-carboxamide, Histone-lysine N-methyltransferase EZH2,Histone-lysine N-methyltransferase EZH2,Histone-lysine N-methyltransferase EZH2, Jarid2 K116me3, ... | | Authors: | Zhang, Y, Justin, N, Chen, S, Wilson, J, Gamblin, S. | | Deposit date: | 2016-08-22 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | Identification of (R)-N-((4-Methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl)-2-methyl-1-(1-(1-(2,2,2-trifluoroethyl)piperidin-4-yl)ethyl)-1H-indole-3-carboxamide (CPI-1205), a Potent and Selective Inhibitor of Histone Methyltransferase EZH2, Suitable for Phase I Clinical Trials for B-Cell Lymphomas.
J. Med. Chem., 59, 2016
|
|
4JND
 
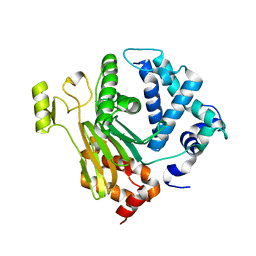 | | Structure of a C.elegans sex determining protein | | Descriptor: | Ca(2+)/calmodulin-dependent protein kinase phosphatase, MAGNESIUM ION | | Authors: | Feng, Y, Zhang, Y, Ge, J, Yang, M. | | Deposit date: | 2013-03-15 | | Release date: | 2013-06-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.652 Å) | | Cite: | Structural insight into Caenorhabditis elegans sex-determining protein FEM-2.
J.Biol.Chem., 288, 2013
|
|
4K7F
 
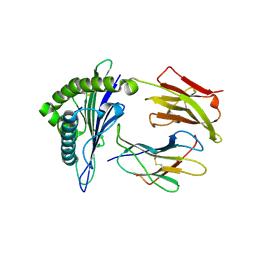 | | Newly identified epitope V60 from HBV core protein complexed with HLA-A*0201 | | Descriptor: | Beta-2-microglobulin, Core protein, HLA class I histocompatibility antigen, ... | | Authors: | Meng, S.D, Zhang, Y, Wu, Y, Qi, J.X. | | Deposit date: | 2013-04-17 | | Release date: | 2013-06-05 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The L60V variation in hepatitis B virus core protein elicits new epitope-specific cytotoxic T lymphocytes and enhances viral replication.
J.Virol., 87, 2013
|
|
6E86
 
 | |
1R0V
 
 | |
1R11
 
 | |
6KQD
 
 | | Thermus thermophilus initial transcription complex comprising sigma A and 5'-OH RNA of 3 nt | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, DNA (5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*CP*AP*GP*GP*G)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G*)-3'), ... | | Authors: | Zhang, Y, Li, L, Ebright, R.H. | | Deposit date: | 2019-08-17 | | Release date: | 2020-03-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6KQE
 
 | | Thermus thermophilus initial transcription complex comprising sigma A and 5'-OH RNA of 4 nt | | Descriptor: | DNA (5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G*)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Zhang, Y, Li, L, Ebright, R.H. | | Deposit date: | 2019-08-17 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6KQG
 
 | | Thermus thermophilus initial transcription complex comprising sigma A and 5'-OH RNA of 6 nt | | Descriptor: | DNA (5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G*)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Zhang, Y, Li, L, Ebright, R.H. | | Deposit date: | 2019-08-17 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.783 Å) | | Cite: | RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6KQM
 
 | | Thermus thermophilus initial transcription complex comprising sigma A and 5'-triphosphate RNA of 5 nt | | Descriptor: | DNA (5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G*)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Zhang, Y, Li, L, Ebright, R.H. | | Deposit date: | 2019-08-18 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.197 Å) | | Cite: | RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
