1XRE
 
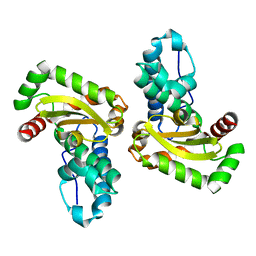 | | Crystal Structure of SodA-2 (BA5696) from Bacillus anthracis at 1.8A Resolution. | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase | | Authors: | Boucher, I.W, Levdikov, V.M, Blagova, E.V, Fogg, M.J, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S. | | Deposit date: | 2004-10-14 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of two superoxide dismutases from Bacillus anthracis reveal a novel active centre.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1XE3
 
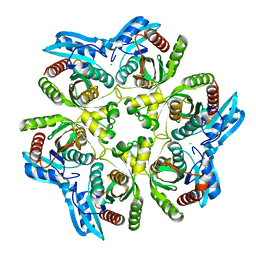 | | Crystal Structure of purine nucleoside phosphorylase DeoD from Bacillus anthracis | | Descriptor: | CHLORIDE ION, purine nucleoside phosphorylase | | Authors: | Grenha, R, Levdikov, V.M, Fogg, M, Blagova, E.V, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-09-09 | | Release date: | 2004-10-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure of purine nucleoside phosphorylase (DeoD) from Bacillus anthracis.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1XP3
 
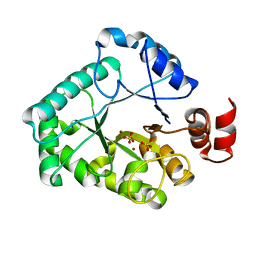 | | Crystal Structure of Endonuclease IV (BA4508) from Bacillus anthracis at 2.57A Resolution. | | Descriptor: | SULFATE ION, ZINC ION, endonuclease IV | | Authors: | Fogg, M.J, Levdikov, V.M, Blagova, E.V, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-10-08 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Crystal Structure of Endonuclease IV (BA4508) from Bacillus anthracis at 2.57A Resolution.
To be Published
|
|
1XKY
 
 | | Crystal Structure of Dihydrodipicolinate Synthase DapA-2 (BA3935) from Bacillus Anthracis at 1.94A Resolution. | | Descriptor: | POTASSIUM ION, dihydrodipicolinate synthase | | Authors: | Levdikov, V, Blagova, E, Fogg, M.J, Brannigan, J.A, Milioti, N, Wilkinson, A.J, Wilson, K.S. | | Deposit date: | 2004-09-30 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of dihydrodipicolinate synthase (BA3935) from Bacillus anthracis at 1.94 A resolution
Proteins, 62, 2006
|
|
1XMP
 
 | | Crystal Structure of PurE (BA0288) from Bacillus anthracis at 1.8 Resolution | | Descriptor: | phosphoribosylaminoimidazole carboxylase | | Authors: | Boyle, M.P, Kalliomaa, A.K, Levdikov, V, Blagova, E, Fogg, M.J, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S. | | Deposit date: | 2004-10-04 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of PurE (BA0288) from Bacillus anthracis at 1.8 A resolution
Proteins, 61, 2005
|
|
1XT8
 
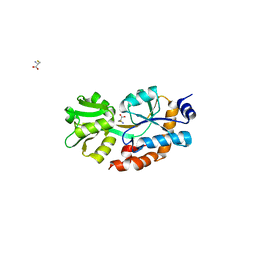 | | Crystal Structure of Cysteine-Binding Protein from Campylobacter jejuni at 2.0 A Resolution | | Descriptor: | CYSTEINE, GLYCEROL, putative amino-acid transporter periplasmic solute-binding protein | | Authors: | Muller, A, Thomas, G.H, Horler, R, Brannigan, J.A, Blagova, E, Levdikov, V.M, Fogg, M.J, Wilson, K.S, Wilkinson, A.J, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-10-21 | | Release date: | 2005-08-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An ATP-binding cassette-type cysteine transporter in Campylobacter jejuni inferred from the structure of an extracytoplasmic solute receptor protein.
Mol.Microbiol., 57, 2005
|
|
1XUQ
 
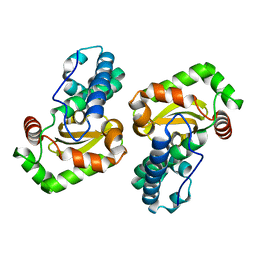 | | Crystal Structure of SodA-1 (BA4499) from Bacillus anthracis at 1.8A Resolution. | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase | | Authors: | Boucher, I.W, Levdikov, V.M, Blagova, E.V, Fogg, M.J, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S. | | Deposit date: | 2004-10-26 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of two superoxide dismutases from Bacillus anthracis reveal a novel active centre.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1QH6
 
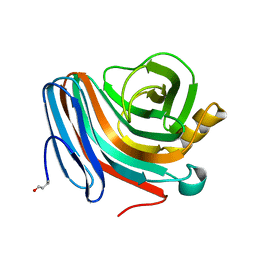 | | CATALYSIS AND SPECIFICITY IN ENZYMATIC GLYCOSIDE HYDROLASES: A 2,5B CONFORMATION FOR THE GLYCOSYL-ENZYME INTERMIDIATE REVEALED BY THE STRUCTURE OF THE BACILLUS AGARADHAERENS FAMILY 11 XYLANASE | | Descriptor: | XYLANASE, beta-D-xylopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-xylopyranose | | Authors: | Sabini, E, Sulzenbacher, G, Dauter, M, Dauter, Z, Jorgensen, P.L, Schulein, M, Dupont, C, Davies, G.J, Wilson, K.S. | | Deposit date: | 1999-05-11 | | Release date: | 2000-05-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catalysis and specificity in enzymatic glycoside hydrolysis: a 2,5B conformation for the glycosyl-enzyme intermediate revealed by the structure of the Bacillus agaradhaerens family 11 xylanase.
Chem.Biol., 6, 1999
|
|
1QNS
 
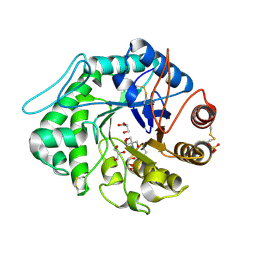 | | The 3-D structure of a Trichoderma reesei b-mannanase from glycoside hydrolase family 5 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDO-1,4-B-D-MANNANASE, ... | | Authors: | Sabini, E, Schubert, H, Murshudov, G, Wilson, K.S, Siika-Aho, M, Penttila, M. | | Deposit date: | 1999-10-20 | | Release date: | 2000-10-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Three-Dimensional Structure of a Trichoderma Reesei Beta-Mannanase from Glycoside Hydrolase Family 5
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1QBA
 
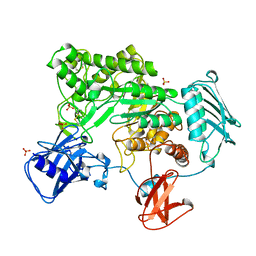 | | BACTERIAL CHITOBIASE, GLYCOSYL HYDROLASE FAMILY 20 | | Descriptor: | CHITOBIASE, SULFATE ION | | Authors: | Tews, I, Perrakis, A, Oppenheim, A, Dauter, Z, Wilson, K.S, Vorgias, C.E. | | Deposit date: | 1996-06-06 | | Release date: | 1997-01-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Bacterial chitobiase structure provides insight into catalytic mechanism and the basis of Tay-Sachs disease.
Nat.Struct.Biol., 3, 1996
|
|
1QH7
 
 | | CATALYSIS AND SPECIFICITY IN ENZYMATIC GLYCOSIDE HYDROLASES: A 2,5B CONFORMATION FOR THE GLYCOSYL-ENZYME INTERMIDIATE REVEALED BY THE STRUCTURE OF THE BACILLUS AGARADHAERENS FAMILY 11 XYLANASE | | Descriptor: | XYLANASE, beta-D-xylopyranose | | Authors: | Sabini, E, Sulzenbacher, G, Dauter, M, Dauter, Z, Jorgensen, P.L, Schulein, M, Dupont, C, Davies, G.J, Wilson, K.S. | | Deposit date: | 1999-05-11 | | Release date: | 2000-05-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Catalysis and specificity in enzymatic glycoside hydrolysis: a 2,5B conformation for the glycosyl-enzyme intermediate revealed by the structure of the Bacillus agaradhaerens family 11 xylanase.
Chem.Biol., 6, 1999
|
|
1QHP
 
 | | FIVE-DOMAIN ALPHA-AMYLASE FROM BACILLUS STEAROTHERMOPHILUS, MALTOSE COMPLEX | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, SULFATE ION, ... | | Authors: | Dauter, Z, Dauter, M, Brzozowski, A.M, Christensen, S, Borchert, T.V, Beier, L, Wilson, K.S, Davies, G.J. | | Deposit date: | 1999-05-25 | | Release date: | 2000-05-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray structure of Novamyl, the five-domain "maltogenic" alpha-amylase from Bacillus stearothermophilus: maltose and acarbose complexes at 1.7A resolution.
Biochemistry, 38, 1999
|
|
1QNR
 
 | | The 3-D structure of a Trichoderma reesei b-mannanase from glycoside hydrolase family 5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDO-1,4-B-D-MANNANASE, GLYCEROL, ... | | Authors: | Sabini, E, Schubert, H, Murshudov, G, Wilson, K.S, Siika-Aho, M, Penttila, M. | | Deposit date: | 1999-10-20 | | Release date: | 2000-10-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Three-Dimensional Structure of a Trichoderma Reesei Beta-Mannanase from Glycoside Hydrolase Family 5.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1QNQ
 
 | | The 3-D structure of a Trichoderma reesei b-mannanase from glycoside hydrolase family 5 | | Descriptor: | 2,2':6',2''-TERPYRIDINE PLATINUM(II) Chloride, 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDO-1,4-B-D-MANNANASE, ... | | Authors: | Sabini, E, Schubert, H, Murshudov, G, Wilson, K.S, Siika-Aho, M, Penttila, M. | | Deposit date: | 1999-10-20 | | Release date: | 2000-10-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Three-Dimensional Structure of a Trichoderma Reesei Beta-Mannanase from Glycoside Hydrolase Family 5.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1OTG
 
 | | 5-CARBOXYMETHYL-2-HYDROXYMUCONATE ISOMERASE | | Descriptor: | 5-CARBOXYMETHYL-2-HYDROXYMUCONATE ISOMERASE, SULFATE ION | | Authors: | Subramanya, H.S, Roper, D.I, Dauter, Z, Dodson, E.J, Davies, G.J, Wilson, K.S, Wigley, D.B. | | Deposit date: | 1995-11-09 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Enzymatic ketonization of 2-hydroxymuconate: specificity and mechanism investigated by the crystal structures of two isomerases.
Biochemistry, 35, 1996
|
|
1OGK
 
 | | The crystal structure of Trypanosoma cruzi dUTPase in complex with dUDP | | Descriptor: | DEOXYURIDINE TRIPHOSPHATASE, DEOXYURIDINE-5'-DIPHOSPHATE | | Authors: | Harkiolaki, M, Dodson, E.J, Bernier-Villamor, V, Turkenburg, J.P, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2003-05-07 | | Release date: | 2004-01-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The Crystal Structure of Trypanosoma Cruzi Dutpase Reveals a Novel Dutp/Dudp Binding Fold
Structure, 12, 2004
|
|
1QHO
 
 | | FIVE-DOMAIN ALPHA-AMYLASE FROM BACILLUS STEAROTHERMOPHILUS, MALTOSE/ACARBOSE COMPLEX | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, SULFATE ION, ... | | Authors: | Dauter, Z, Dauter, M, Brzozowski, A.M, Christensen, S, Borchert, T.V, Beier, L, Wilson, K.S, Davies, G.J. | | Deposit date: | 1999-05-25 | | Release date: | 2000-05-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray structure of Novamyl, the five-domain "maltogenic" alpha-amylase from Bacillus stearothermophilus: maltose and acarbose complexes at 1.7A resolution.
Biochemistry, 38, 1999
|
|
1QGO
 
 | | ANAEROBIC COBALT CHELATASE IN COBALAMIN BIOSYNTHESIS FROM SALMONELLA TYPHIMURIUM | | Descriptor: | ANAEROBIC COBALAMIN BIOSYNTHETIC COBALT CHELATASE, SULFATE ION | | Authors: | Schubert, H.L, Raux, E, Warren, M.J, Wilson, K.S. | | Deposit date: | 1999-05-03 | | Release date: | 1999-09-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Common chelatase design in the branched tetrapyrrole pathways of heme and anaerobic cobalamin synthesis.
Biochemistry, 38, 1999
|
|
1QNP
 
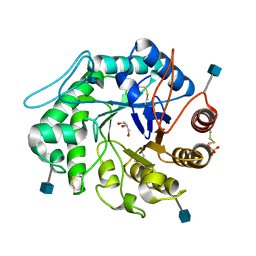 | | The 3-D structure of a Trichoderma reesei b-mannanase from glycoside hydrolase family 5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDO-1,4-B-D-MANNANASE, GLYCEROL, ... | | Authors: | Sabini, E, Schubert, H, Murshudov, G, Wilson, K.S, Siika-Aho, M, Penttila, M. | | Deposit date: | 1999-10-20 | | Release date: | 2000-10-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Three-Dimensional Structure of a Trichoderma Reesei Beta-Mannanase from Glycoside Hydrolase Family 5.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1QBB
 
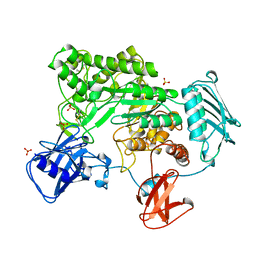 | | BACTERIAL CHITOBIASE COMPLEXED WITH CHITOBIOSE (DINAG) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITOBIASE, SULFATE ION | | Authors: | Tews, I, Perrakis, A, Oppenheim, A, Dauter, Z, Wilson, K.S, Vorgias, C.E. | | Deposit date: | 1996-06-07 | | Release date: | 1997-02-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bacterial chitobiase structure provides insight into catalytic mechanism and the basis of Tay-Sachs disease.
Nat.Struct.Biol., 3, 1996
|
|
1PY3
 
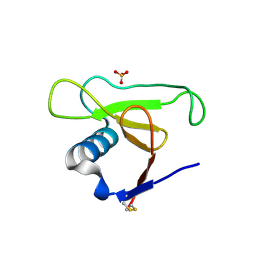 | | Crystal structure of Ribonuclease Sa2 | | Descriptor: | SULFATE ION, ribonuclease | | Authors: | Sevcik, J, Dauter, Z, Wilson, K.S. | | Deposit date: | 2003-07-08 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure reveals two alternative conformations in the active site of ribonuclease Sa2.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1PYL
 
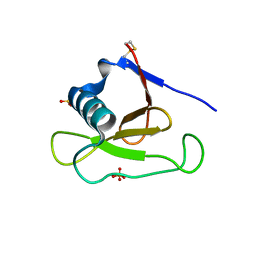 | | Crystal structure of Ribonuclease Sa2 | | Descriptor: | SULFATE ION, ribonuclease | | Authors: | Sevcik, J, Dauter, Z, Wilson, K.S. | | Deposit date: | 2003-07-09 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.507 Å) | | Cite: | Crystal structure reveals two alternative conformations in the active site of ribonuclease Sa2.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1S4D
 
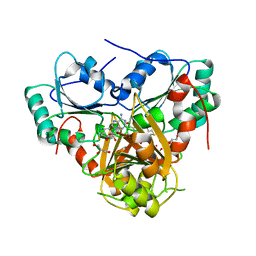 | | Crystal Structure Analysis of the S-adenosyl-L-methionine dependent uroporphyrinogen-III C-methyltransferase SUMT | | Descriptor: | GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, Uroporphyrin-III C-methyltransferase | | Authors: | Vevodova, J, Graham, R.M, Raux, E, Schubert, H.L, Roper, D.I, Brindley, A.A, Scott, A.I, Roessner, C.A, Stamford, N.P.J, Stroupe, M.E, Getzoff, E.D, Warren, M.J, Wilson, K.S. | | Deposit date: | 2004-01-16 | | Release date: | 2004-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure/Function Studies on a S-Adenosyl-l-methionine-dependent Uroporphyrinogen III C Methyltransferase (SUMT), a Key Regulatory Enzyme of Tetrapyrrole Biosynthesis
J.Mol.Biol., 344, 2004
|
|
1PSP
 
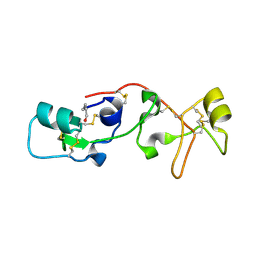 | | PANCREATIC SPASMOLYTIC POLYPEPTIDE: FIRST THREE-DIMENSIONAL STRUCTURE OF A MEMBER OF THE MAMMALIAN TREFOIL FAMILY OF PEPTIDES | | Descriptor: | PANCREATIC SPASMOLYTIC POLYPEPTIDE | | Authors: | Gajhede, M, Petersen, T.N, Henriksen, A, Petersen, J.F.W, Dauter, Z, Wilson, K.S, Thim, L. | | Deposit date: | 1994-01-05 | | Release date: | 1994-04-30 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Pancreatic spasmolytic polypeptide: first three-dimensional structure of a member of the mammalian trefoil family of peptides.
Structure, 1, 1993
|
|
1RB9
 
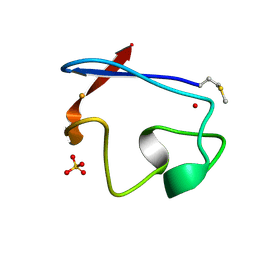 | | RUBREDOXIN FROM DESULFOVIBRIO VULGARIS REFINED ANISOTROPICALLY AT 0.92 ANGSTROMS RESOLUTION | | Descriptor: | FE (II) ION, RUBREDOXIN, SULFATE ION | | Authors: | Dauter, Z, Butterworth, S, Sieker, L.C, Sheldrick, G, Wilson, K.S. | | Deposit date: | 1997-12-21 | | Release date: | 1999-02-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Anisotropic Refinement of Rubredoxin from Desulfovibrio Vulgaris
To be Published
|
|
