4PII
 
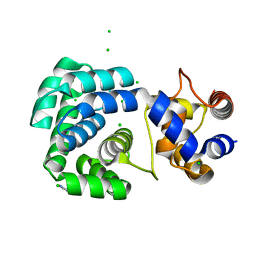 | | Crystal structure of hypothetical protein PF0907 from pyrococcus furiosus solved by sulfur SAD using Swiss light source data | | Descriptor: | CHLORIDE ION, IMIDAZOLE, N-glycosylase/DNA lyase | | Authors: | Weinert, T, Waltersperger, S, Olieric, V, Panepucci, E, Chen, L, Rose, J.P, Wang, M, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2014-05-08 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
1KVK
 
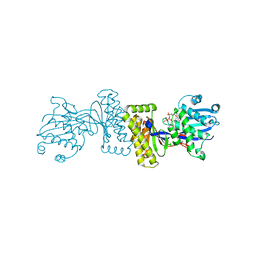 | | The Structure of Binary complex between a Mammalian Mevalonate Kinase and ATP: Insights into the Reaction Mechanism and Human Inherited Disease | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, mevalonate kinase | | Authors: | Fu, Z, Wang, M, Potter, D, Mizioko, H.M, Kim, J.J. | | Deposit date: | 2002-01-26 | | Release date: | 2002-03-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Structure of a Binary complex
between a Mammalian Mevalonate Kinase and ATP: Insights into the
Reaction Mechanism and Human Inherited Disease
J.Biol.Chem., 277, 2002
|
|
4A3Y
 
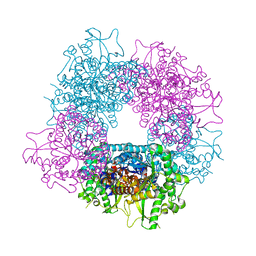 | | Crystal structure of Raucaffricine glucosidase from ajmaline biosynthesis pathway | | Descriptor: | GLYCEROL, RAUCAFFRICINE-O-BETA-D-GLUCOSIDASE, SULFATE ION | | Authors: | Xia, L, Ruppert, M, Wang, M, Panjikar, S, Barleben, L, Rajendran, C, Lin, H, Stoeckigt, J. | | Deposit date: | 2011-10-06 | | Release date: | 2012-08-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of Alkaloid Biosynthetic Glucosidases Decode Substrate Specificity.
Acs Chem.Biol., 7, 2012
|
|
5KWB
 
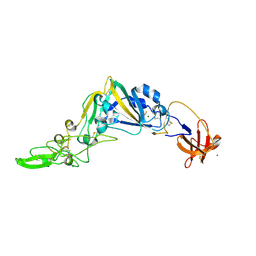 | | Crystal Structure of the Receptor Binding Domain of the Spike Glycoprotein of Human Betacoronavirus HKU1 (HKU1 1A-CTD, 1.9 angstrom, molecular replacement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Spike glycoprotein, ... | | Authors: | Guan, H, Wojdyla, J.A, Wang, M, Cui, S. | | Deposit date: | 2016-07-17 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of the receptor binding domain of the spike glycoprotein of human betacoronavirus HKU1
Nat Commun, 8, 2017
|
|
3R45
 
 | | Structure of a CENP-A-Histone H4 Heterodimer in complex with chaperone HJURP | | Descriptor: | GLYCEROL, Histone H3-like centromeric protein A, Histone H4, ... | | Authors: | Hu, H, Liu, Y, Wang, M, Fang, J, Huang, H, Yang, N, Li, Y, Wang, J, Yao, X, Shi, Y, Li, G, Xu, R.M. | | Deposit date: | 2011-03-17 | | Release date: | 2011-04-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP
Genes Dev., 25, 2011
|
|
8XJ3
 
 | |
6KHX
 
 | | Crystal structure of Prx from Akkermansia muciniphila | | Descriptor: | CALCIUM ION, Peroxiredoxin | | Authors: | Li, M, Wang, J, Xu, W, Wang, Y, Zhang, M, Wang, M. | | Deposit date: | 2019-07-16 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal structure of Akkermansia muciniphila peroxiredoxin reveals a novel regulatory mechanism of typical 2-Cys Prxs by a distinct loop.
Febs Lett., 594, 2020
|
|
2AKJ
 
 | | Structure of spinach nitrite reductase | | Descriptor: | Ferredoxin--nitrite reductase, chloroplast, IRON/SULFUR CLUSTER, ... | | Authors: | Swamy, U, Wang, M, Tripathy, J.N, Kim, S.-K, Hirasawa, M, Knaff, D.B, Allen, J.P. | | Deposit date: | 2005-08-03 | | Release date: | 2006-01-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Spinach Nitrite Reductase: Implications for Multi-electron Reactions by the Iron-Sulfur:Siroheme Cofactor
Biochemistry, 44, 2005
|
|
7DJZ
 
 | | Crystal structure of SARS-CoV-2 Spike RBD in complex with MW01 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, MW01 heavy chain, ... | | Authors: | Wang, J, Jiao, S, Wang, R, Zhang, J, Zhang, M, Wang, M. | | Deposit date: | 2020-11-22 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Antibody-dependent enhancement (ADE) of SARS-CoV-2 pseudoviral infection requires Fc gamma RIIB and virus-antibody complex with bivalent interaction.
Commun Biol, 5, 2022
|
|
7DK0
 
 | | Crystal structure of SARS-CoV-2 Spike RBD in complex with MW05 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MW05 heavy chain, MW05 light chain, ... | | Authors: | Wang, J, Jiao, S, Wang, R, Zhang, J, Zhang, M, Wang, M. | | Deposit date: | 2020-11-22 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.199 Å) | | Cite: | Antibody-dependent enhancement (ADE) of SARS-CoV-2 pseudoviral infection requires Fc gamma RIIB and virus-antibody complex with bivalent interaction.
Commun Biol, 5, 2022
|
|
5YEG
 
 | | Crystal structure of CTCF ZFs4-8-Hs5-1a complex | | Descriptor: | DNA (5'-D(*AP*CP*TP*TP*TP*AP*AP*CP*CP*AP*GP*CP*AP*GP*AP*GP*GP*GP*CP*G)-3'), DNA (5'-D(*TP*CP*GP*CP*CP*CP*TP*CP*TP*GP*CP*TP*GP*GP*TP*TP*AP*AP*AP*G)-3'), Transcriptional repressor CTCF, ... | | Authors: | Yin, M, Wang, J, Wang, M, Li, X. | | Deposit date: | 2017-09-17 | | Release date: | 2017-11-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular mechanism of directional CTCF recognition of a diverse range of genomic sites
Cell Res., 27, 2017
|
|
4RLM
 
 | | Hen egg-white lysozyme solved from serial crystallography at a synchrotron source, data processed with CrystFEL | | Descriptor: | Lysozyme C | | Authors: | Botha, S, Nass, K, Barends, T, Kabsch, W, Latz, B, Dworkowski, F, Foucar, L, Panepucci, E, Wang, M, Shoeman, R, Schlichting, I, Doak, R.B. | | Deposit date: | 2014-10-17 | | Release date: | 2015-02-25 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Room-temperature serial crystallography at synchrotron X-ray sources using slowly flowing free-standing high-viscosity microstreams.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3GZJ
 
 | | Crystal Structure of Polyneuridine Aldehyde Esterase Complexed with 16-epi-Vellosimine | | Descriptor: | 16-epi-Vellosimine, Polyneuridine-aldehyde esterase | | Authors: | Yang, L, Hill, M, Wang, M, Panjikar, S, Stoeckigt, J. | | Deposit date: | 2009-04-07 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural basis and enzymatic mechanism of the biosynthesis of C9- from C10-monoterpenoid indole alkaloids
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
5MKK
 
 | | Crystal structure of the heterodimeric ABC transporter TmrAB, a homolog of the antigen translocation complex TAP | | Descriptor: | Multidrug resistance ABC transporter ATP-binding and permease protein, SULFATE ION | | Authors: | Noell, A, Thomas, C, Tomasiak, T.M, Olieric, V, Wang, M, Diederichs, K, Stroud, R.M, Pos, K.M, Tampe, R. | | Deposit date: | 2016-12-05 | | Release date: | 2017-01-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and mechanistic basis of a functional homolog of the antigen transporter TAP.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5YEL
 
 | | Crystal structure of CTCF ZFs6-11-gb7CSE | | Descriptor: | DNA (26-MER), Transcriptional repressor CTCF, ZINC ION | | Authors: | Yin, M, Wang, J, Wang, M, Li, X, Wang, Y. | | Deposit date: | 2017-09-18 | | Release date: | 2017-11-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Molecular mechanism of directional CTCF recognition of a diverse range of genomic sites
Cell Res., 27, 2017
|
|
5YEH
 
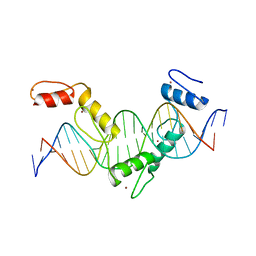 | | Crystal structure of CTCF ZFs4-8-eCBS | | Descriptor: | DNA (5'-D(*AP*CP*GP*GP*TP*TP*TP*CP*CP*GP*CP*TP*AP*GP*AP*GP*GP*GP*CP*G)-3'), DNA (5'-D(*TP*CP*GP*CP*CP*CP*TP*CP*TP*AP*GP*CP*GP*GP*AP*AP*AP*CP*CP*G)-3'), Transcriptional repressor CTCF, ... | | Authors: | Yin, M, Wang, J, Wang, M, Li, X, Wang, Y. | | Deposit date: | 2017-09-17 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.328 Å) | | Cite: | Molecular mechanism of directional CTCF recognition of a diverse range of genomic sites
Cell Res., 27, 2017
|
|
5YEF
 
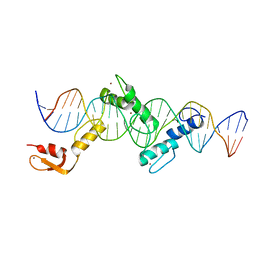 | | Crystal structure of CTCF ZFs2-8-Hs5-1aE | | Descriptor: | DNA (27-MER), Transcriptional repressor CTCF, ZINC ION | | Authors: | Yin, M, Wang, J, Wang, M, Li, X, Wang, Y. | | Deposit date: | 2017-09-17 | | Release date: | 2017-11-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.807 Å) | | Cite: | Molecular mechanism of directional CTCF recognition of a diverse range of genomic sites
Cell Res., 27, 2017
|
|
4RLN
 
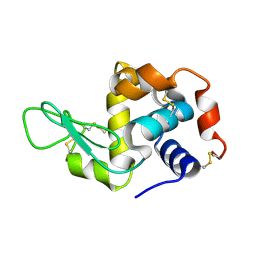 | | Hen egg-white lysozyme solved from serial crystallography at a synchrotron source, data processed with nXDS | | Descriptor: | Lysozyme C | | Authors: | Botha, S, Nass, K, Barends, T, Kabsch, W, Latz, B, Dworkowski, F, Foucar, L, Panepucci, E, Wang, M, Shoeman, R, Schlichting, I, Doak, R.B. | | Deposit date: | 2014-10-17 | | Release date: | 2015-02-25 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Room-temperature serial crystallography at synchrotron X-ray sources using slowly flowing free-standing high-viscosity microstreams.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6OM3
 
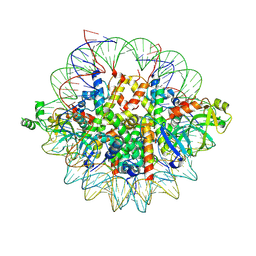 | | Crystal structure of the Orc1 BAH domain in complex with a nucleosome core particle | | Descriptor: | DNA (146-MER), DNA (147-MER), Histone H2A, ... | | Authors: | De Ioannes, P.E, Wang, M, Armache, K.-J. | | Deposit date: | 2019-04-18 | | Release date: | 2019-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure and function of the Orc1 BAH-nucleosome complex.
Nat Commun, 10, 2019
|
|
4EQ3
 
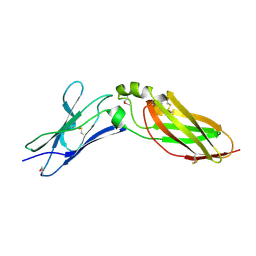 | | Crystal Structure Analysis of Selenomethionine (Se-Met) Substituted Chicken Interferon Gamma Receptor Alpha Chain | | Descriptor: | Interferon gamma receptor 1 | | Authors: | Ping, Z, Qi, J, Lu, G, Shi, Y, Wang, X, Gao, G.F, Wang, M. | | Deposit date: | 2012-04-18 | | Release date: | 2013-04-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal structure of the interferon gamma receptor alpha chain from chicken reveals an undetected extra helix compared with the human counterparts.
J.Interferon Cytokine Res., 34, 2014
|
|
2WFL
 
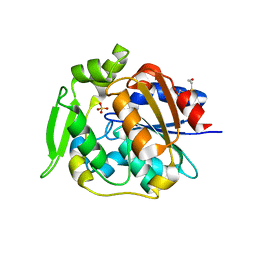 | | Crystal structure of polyneuridine aldehyde esterase | | Descriptor: | POLYNEURIDINE-ALDEHYDE ESTERASE, SULFATE ION | | Authors: | Yang, L, Hill, M, Panjikar, S, Wang, M, Stoeckigt, J. | | Deposit date: | 2009-04-08 | | Release date: | 2009-08-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis and Enzymatic Mechanism of the Biosynthesis of C9- from C10-Monoterpenoid Indole Alkaloids.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
2WFM
 
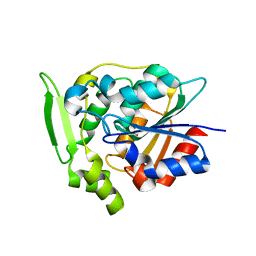 | | Crystal structure of polyneuridine aldehyde esterase mutant (H244A) | | Descriptor: | POLYNEURIDINE ALDEHYDE ESTERASE | | Authors: | Yang, L, Hill, M, Panjikar, S, Wang, M, Stoeckigt, J. | | Deposit date: | 2009-04-08 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis and Enzymatic Mechanism of the Biosynthesis of C9- from C10-Monoterpenoid Indole Alkaloids.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
4EQ2
 
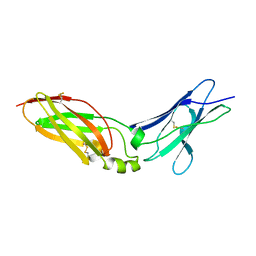 | | Crystal Structure Analysis of Chicken Interferon Gamma Receptor Alpha Chain | | Descriptor: | Interferon gamma receptor 1 | | Authors: | Ping, Z, Qi, J, Lu, G, Shi, Y, Wang, X, Gao, G.F, Wang, M. | | Deposit date: | 2012-04-18 | | Release date: | 2013-04-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Crystal structure of the interferon gamma receptor alpha chain from chicken reveals an undetected extra helix compared with the human counterparts.
J.Interferon Cytokine Res., 34, 2014
|
|
4R8U
 
 | | S-SAD structure of DINB-DNA Complex | | Descriptor: | 5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]thymidine, DNA, DNA polymerase IV, ... | | Authors: | Kottur, J, Nair, D.T, Weinert, T, Oligeric, V, Wang, M. | | Deposit date: | 2014-09-03 | | Release date: | 2015-01-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination
Nat.Methods, 12, 2015
|
|
4R8T
 
 | | Structure of JEV protease | | Descriptor: | CHLORIDE ION, NS3, Serine protease subunit NS2B | | Authors: | Nair, D.T, Weinert, T, Wang, M, Olieric, V. | | Deposit date: | 2014-09-03 | | Release date: | 2014-12-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.133 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
