7S7D
 
 | | STRUCTURE OF HLA-B*07:02 IN COMPLEX WITH SYNTHETIC SULFO-MLL PEPTIDE ANALOG | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Patskovska, L, Patskovsky, Y, Nyovanie, S, Natarajan, A, Joshi, B, Morin, B, Brittsan, C, Huber, O, Gordon, S, Michelet, X, Schmitzberger, F, Stein, R, Findeis, M, Hurwitz, A, Van Dijk, M, Buell, J, Underwood, D, Krogsgaard, M. | | Deposit date: | 2021-09-15 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Molecular mechanism of phosphopeptide neoantigen immunogenicity.
Nat Commun, 14, 2023
|
|
7S8E
 
 | | STRUCTURE OF HLA-B*07:02 IN COMPLEX WITH MLL(747-755) PHOSPHOPEPTIDE AND BOUND GLYCEROL | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Patskovsky, Y, Nyovanie, S, Patskovska, L, Natarajan, A, Joshi, B, Morin, B, Brittsan, C, Huber, O, Gordon, S, Michelet, X, Schmitzberger, F, Stein, R, Findeis, M, Hurwitz, A, Van Dijk, M, Buell, J, Underwood, D, Krogsgaard, M. | | Deposit date: | 2021-09-17 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular mechanism of phosphopeptide neoantigen immunogenicity.
Nat Commun, 14, 2023
|
|
7RZJ
 
 | | CRYSTAL STRUCTURE OF HLA-B*07:02 IN COMPLEX WITH MLL(747-755) PHOSPHOPEPTIDE | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-7 alpha chain, ... | | Authors: | Patskovsky, Y, Patskovska, L, Nyovanie, S, Natarajan, A, Joshi, B, Morin, B, Brittsan, C, Huber, O, Gordon, S, Michelet, X, Schmitzberger, F, Stein, R, Findeis, M, Hurwitz, A, Van Dijk, M, Buell, J, Underwood, D, Krogsgaard, M. | | Deposit date: | 2021-08-27 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular mechanism of phosphopeptide neoantigen immunogenicity.
Nat Commun, 14, 2023
|
|
7S8F
 
 | | STRUCTURE OF HLA-B*07:02 IN COMPLEX WITH MLL(747-755) PEPTIDE AND BOUND GLYCEROL | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Patskovsky, Y, Nyovanie, S, Patskovska, L, Natarajan, A, Joshi, B, Morin, B, Brittsan, C, Huber, O, Gordon, S, Michelet, X, Schmitzberger, F, Stein, R, Findeis, M, Hurwitz, A, Van Dijk, M, Buell, J, Underwood, D, Krogsgaard, M. | | Deposit date: | 2021-09-17 | | Release date: | 2022-11-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular mechanism of phosphopeptide neoantigen immunogenicity.
Nat Commun, 14, 2023
|
|
8F11
 
 | | T4 lysozyme with a 2,6-diazaadamantane nitroxide (DZD) spin label | | Descriptor: | 1-[(1r,3r,5r,7r)-6-hydroxy-2,6-diazatricyclo[3.3.1.1~3,7~]decan-2-yl]ethan-1-one, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Wilson, M.A, Madzelan, P, Rajca, A, Stein, R, Yang, Z. | | Deposit date: | 2022-11-04 | | Release date: | 2023-02-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Cucurbit[7]uril Enhances Distance Measurements of Spin-Labeled Proteins.
J.Am.Chem.Soc., 145, 2023
|
|
8C4Y
 
 | | SFX structure of FutA bound to Fe(III) | | Descriptor: | FE (III) ION, Putative iron ABC transporter, substrate binding protein | | Authors: | Bolton, R, Tews, I. | | Deposit date: | 2023-01-05 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A redox switch allows binding of Fe(II) and Fe(III) ions in the cyanobacterial iron-binding protein FutA from Prochlorococcus.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8OGG
 
 | | Crystal structure of FutA after an accumulated dose of 5 kGy | | Descriptor: | FE (III) ION, Putative iron ABC transporter, substrate binding protein | | Authors: | Bolton, R, Tews, I. | | Deposit date: | 2023-03-20 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A redox switch allows binding of Fe(II) and Fe(III) ions in the cyanobacterial iron-binding protein FutA from Prochlorococcus.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8OEM
 
 | | Crystal structure of FutA bound to Fe(II) | | Descriptor: | FE (II) ION, Putative iron ABC transporter, substrate binding protein | | Authors: | Bolton, R, Tews, I. | | Deposit date: | 2023-03-10 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A redox switch allows binding of Fe(II) and Fe(III) ions in the cyanobacterial iron-binding protein FutA from Prochlorococcus.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8OEI
 
 | | SFX structure of FutA after an accumulated dose of 350 kGy | | Descriptor: | FE (III) ION, Putative iron ABC transporter, substrate binding protein | | Authors: | Bolton, R, Tews, I. | | Deposit date: | 2023-03-10 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A redox switch allows binding of Fe(II) and Fe(III) ions in the cyanobacterial iron-binding protein FutA from Prochlorococcus.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
1ZIS
 
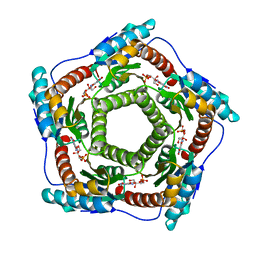 | | Recombinant Lumazine synthase (hexagonal form) | | Descriptor: | 5-NITRO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION | | Authors: | Lopez-Jaramillo, F.J. | | Deposit date: | 2005-04-27 | | Release date: | 2006-10-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of recombinant Lumazine synthase (hexagonal form)
To be Published
|
|
8RK1
 
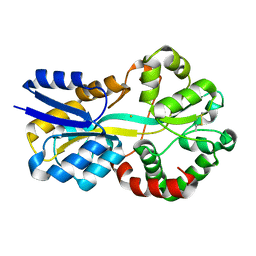 | | Crystal structure of FutA bound to Fe(III) solved by neutron diffraction | | Descriptor: | FE (III) ION, Putative iron ABC transporter, substrate binding protein | | Authors: | Bolton, R, Tews, I. | | Deposit date: | 2023-12-22 | | Release date: | 2024-01-17 | | Last modified: | 2024-10-16 | | Method: | NEUTRON DIFFRACTION (2.095 Å) | | Cite: | A redox switch allows binding of Fe(II) and Fe(III) ions in the cyanobacterial iron-binding protein FutA from Prochlorococcus.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
1GNE
 
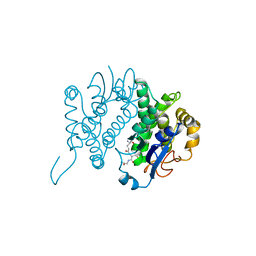 | | THE THREE-DIMENSIONAL STRUCTURE OF GLUTATHIONE S-TRANSFERASE OF SCHISTOSOMA JAPONICUM FUSED WITH A CONSERVED NEUTRALIZING EPITOPE ON GP41 OF HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 | | Descriptor: | GLUTATHIONE, GLUTATHIONE S-TRANSFERASE | | Authors: | Lim, K, Ho, J.X, Keeling, K, Gilliland, G.L, Ji, X, Ruker, F, Carter, D.C. | | Deposit date: | 1994-06-16 | | Release date: | 1994-11-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structure of Schistosoma japonicum glutathione S-transferase fused with a six-amino acid conserved neutralizing epitope of gp41 from HIV.
Protein Sci., 3, 1994
|
|
1ASO
 
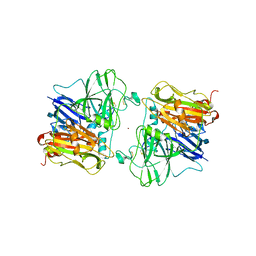 | | X-RAY STRUCTURES AND MECHANISTIC IMPLICATIONS OF THREE FUNCTIONAL DERIVATIVES OF ASCORBATE OXIDASE FROM ZUCCHINI: REDUCED-, PEROXIDE-, AND AZIDE-FORMS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ASCORBATE OXIDASE, COPPER (II) ION, ... | | Authors: | Messerschmidt, A, Luecke, H, Huber, R. | | Deposit date: | 1992-11-25 | | Release date: | 1994-01-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structures and mechanistic implications of three functional derivatives of ascorbate oxidase from zucchini. Reduced, peroxide and azide forms.
J.Mol.Biol., 230, 1993
|
|
1ASQ
 
 | | X-RAY STRUCTURES AND MECHANISTIC IMPLICATIONS OF THREE FUNCTIONAL DERIVATIVES OF ASCORBATE OXIDASE FROM ZUCCHINI: REDUCED-, PEROXIDE-, AND AZIDE-FORMS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ASCORBATE OXIDASE, AZIDE ION, ... | | Authors: | Messerschmidt, A, Luecke, H, Huber, R. | | Deposit date: | 1992-11-25 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | X-ray structures and mechanistic implications of three functional derivatives of ascorbate oxidase from zucchini. Reduced, peroxide and azide forms.
J.Mol.Biol., 230, 1993
|
|
1ASP
 
 | | X-RAY STRUCTURES AND MECHANISTIC IMPLICATIONS OF THREE FUNCTIONAL DERIVATIVES OF ASCORBATE OXIDASE FROM ZUCCHINI: REDUCED-, PEROXIDE-, AND AZIDE-FORMS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ASCORBATE OXIDASE, COPPER (II) ION, ... | | Authors: | Messerschmidt, A, Luecke, H, Huber, R. | | Deposit date: | 1992-11-25 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | X-ray structures and mechanistic implications of three functional derivatives of ascorbate oxidase from zucchini. Reduced, peroxide and azide forms.
J.Mol.Biol., 230, 1993
|
|
8GHN
 
 | |
8GHA
 
 | | Hir3 Arm/Tail, Hir2 WD40, C-terminal Hpc2 | | Descriptor: | Histone promoter control protein 2, Histone transcription regulator 3, Protein HIR2 | | Authors: | Kim, H.J, Murakami, K. | | Deposit date: | 2023-03-09 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Structure of the Hir histone chaperone complex.
Mol.Cell, 84, 2024
|
|
8GHM
 
 | | Hir1 WD40 domains and Asf1/H3/H4 | | Descriptor: | Histone H3, Histone H4, Histone chaperone, ... | | Authors: | Kim, H.J, Murakami, K. | | Deposit date: | 2023-03-10 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structure of the Hir histone chaperone complex.
Mol.Cell, 84, 2024
|
|
8GHL
 
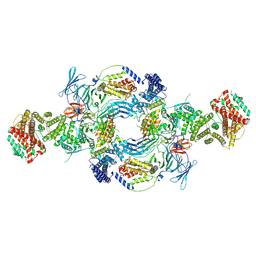 | | the Hir complex core | | Descriptor: | Histone transcription regulator 3, Protein HIR1, Protein HIR2 | | Authors: | Kim, H.J, Murakami, K. | | Deposit date: | 2023-03-10 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structure of the Hir histone chaperone complex.
Mol.Cell, 84, 2024
|
|
3PUP
 
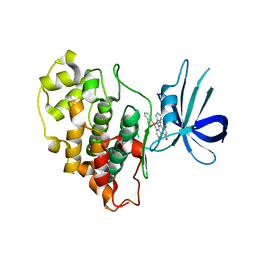 | | Structure of Glycogen Synthase Kinase 3 beta (GSK3B) in complex with a ruthenium octasporine ligand (OS1) | | Descriptor: | Glycogen synthase kinase-3 beta, Ruthenium octasporine | | Authors: | Filippakopoulos, P, Kraling, K, Essen, L.O, Meggers, E, Knapp, S. | | Deposit date: | 2010-12-06 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structurally sophisticated octahedral metal complexes as highly selective protein kinase inhibitors.
J.Am.Chem.Soc., 133, 2011
|
|
7LJE
 
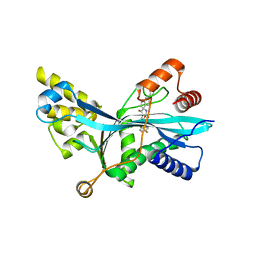 | | Discovery of Spirohydantoins as Selective, Orally Bioavailable Inhibitors of p300/CBP Histone Acetyltransferases | | Descriptor: | 2-[4-[(3'R,4S)-3'-fluoro-1-[2-[(4-fluorophenyl)methyl-[(1S)-2,2,2-trifluoro-1-methyl-ethyl]amino]-2-oxo-ethyl]-2,5-dioxo-spiro[imidazolidine-4,1'-indane]-5'-yl]pyrazol-1-yl]-N-methyl-acetamide, Histone acetyltransferase p300 | | Authors: | Jakob, C.G. | | Deposit date: | 2021-01-29 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.607 Å) | | Cite: | Discovery of spirohydantoins as selective, orally bioavailable inhibitors of p300/CBP histone acetyltransferases.
Bioorg.Med.Chem.Lett., 39, 2021
|
|
3ZXH
 
 | |
7DCJ
 
 | |
7DCU
 
 | | Crystal structure of HSF2 DNA-binding domain in complex with 3-site HSE DNA (21 bp) | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*CP*GP*AP*AP*TP*AP*TP*TP*CP*TP*AP*GP*AP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*TP*CP*TP*AP*GP*AP*AP*TP*AP*TP*TP*CP*GP*CP*GP*G)-3'), Heat shock factor protein 2, ... | | Authors: | Feng, N, Liu, W. | | Deposit date: | 2020-10-27 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of heat shock factor trimers bound to DNA.
Iscience, 24, 2021
|
|
7DCT
 
 | | Crystal structure of HSF1 DNA-binding domain in complex with 3-site HSE DNA (24 bp) | | Descriptor: | DNA (5'-D(*AP*CP*TP*CP*GP*CP*GP*AP*AP*TP*AP*TP*TP*CP*TP*AP*GP*AP*AP*CP*GP*CP*AP*C)-3'), DNA (5'-D(*TP*GP*TP*GP*CP*GP*TP*TP*CP*TP*AP*GP*AP*AP*TP*AP*TP*TP*CP*GP*CP*GP*AP*G)-3'), Heat shock factor protein 1, ... | | Authors: | Feng, N, Liu, W. | | Deposit date: | 2020-10-27 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structures of heat shock factor trimers bound to DNA.
Iscience, 24, 2021
|
|
