7VGK
 
 | | Crystal structure of Lactobacillus rhamnosus 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase KduI | | Descriptor: | 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase | | Authors: | Iwase, H, Oiki, S, Mikami, B, Takase, R, Hashimoto, W. | | Deposit date: | 2021-09-16 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of Lacticaseibacillus 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase KduI in complex with substrate analogs
J.Appl.Glyosci., 2023
|
|
7VER
 
 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in a full open conformation | | Descriptor: | GLYCEROL, SPH1118 | | Authors: | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-09-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
7VEU
 
 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in complex with galacturonic acid | | Descriptor: | GLYCEROL, SPH1118, alpha-D-galactopyranuronic acid | | Authors: | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-09-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.736 Å) | | Cite: | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
7VEW
 
 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in complex with unsaturated trigalacturonic acid | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-09-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
7VEQ
 
 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in an open conformation | | Descriptor: | GLYCEROL, SPH1118 | | Authors: | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-09-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
7VET
 
 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in a closed conformation | | Descriptor: | SPH1118 | | Authors: | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-09-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
7VEV
 
 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in complex with MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, SPH1118 | | Authors: | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-09-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
7YE3
 
 | | Crystal structure of Lactobacillus rhamnosus 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase KduI complexed with MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase, ZINC ION | | Authors: | Yamamoto, Y, Oiki, S, Takase, R, Mikami, B, Hashimoto, W. | | Deposit date: | 2022-07-05 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.553 Å) | | Cite: | Crystal Structures of Lacticaseibacillus 4-Deoxy-L- threo- 5-hexosulose-uronate Ketol-isomerase KduI in Complex with Substrate Analogs.
J Appl Glycosci (1999), 70, 2023
|
|
7WGU
 
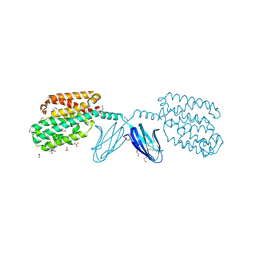 | | Crystal structure of metal-binding protein EfeO from Escherichia coli | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Iron uptake system protein EfeO, ... | | Authors: | Nakatsuji, S, Takase, R, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-12-29 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of EfeB and EfeO in a bacterial siderophore-independent iron transport system
Biochem.Biophys.Res.Commun., 594, 2022
|
|
7YU5
 
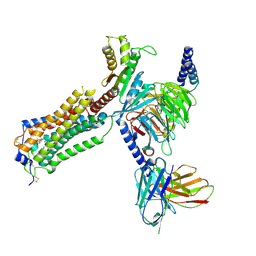 | | Human Lysophosphatidic Acid Receptor 1-Gi complex bound to ONO-0740556, state1 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Akasaka, H, Shihoya, W, Nureki, O. | | Deposit date: | 2022-08-16 | | Release date: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the active G i -coupled human lysophosphatidic acid receptor 1 complexed with a potent agonist.
Nat Commun, 13, 2022
|
|
7YU6
 
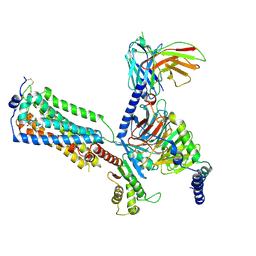 | | Human Lysophosphatidic Acid Receptor 1-Gi complex bound to ONO-0740556, state2 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Akasaka, H, Shihoya, W, Nureki, O. | | Deposit date: | 2022-08-16 | | Release date: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the active G i -coupled human lysophosphatidic acid receptor 1 complexed with a potent agonist.
Nat Commun, 13, 2022
|
|
7YU3
 
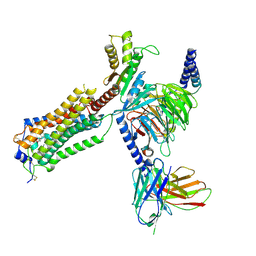 | | Human Lysophosphatidic Acid Receptor 1-Gi complex bound to ONO-0740556 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Akasaka, H, Shihoya, W, Nureki, O. | | Deposit date: | 2022-08-16 | | Release date: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the active G i -coupled human lysophosphatidic acid receptor 1 complexed with a potent agonist.
Nat Commun, 13, 2022
|
|
7YU8
 
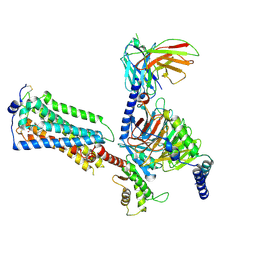 | | Human Lysophosphatidic Acid Receptor 1-Gi complex bound to ONO-0740556, state4 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Akasaka, H, Shihoya, W, Nureki, O. | | Deposit date: | 2022-08-16 | | Release date: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure of the active G i -coupled human lysophosphatidic acid receptor 1 complexed with a potent agonist.
Nat Commun, 13, 2022
|
|
7YU4
 
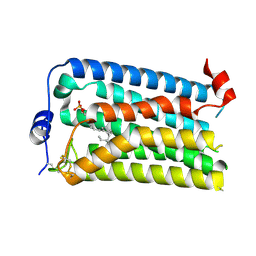 | |
7YU7
 
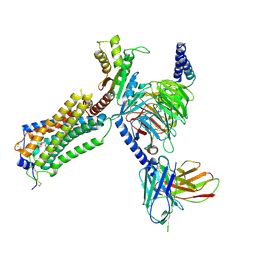 | | Human Lysophosphatidic Acid Receptor 1-Gi complex bound to ONO-0740556, state3 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Akasaka, H, Shihoya, W, Nureki, O. | | Deposit date: | 2022-08-16 | | Release date: | 2022-10-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure of the active G i -coupled human lysophosphatidic acid receptor 1 complexed with a potent agonist.
Nat Commun, 13, 2022
|
|
3VLW
 
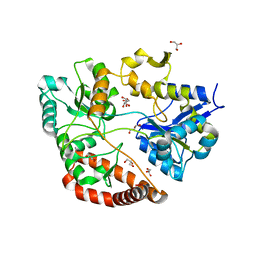 | | Crystal structure of Sphingomonas sp. A1 alginate-binding protein AlgQ1 in complex with mannuronate-guluronate disaccharide | | Descriptor: | AlgQ1, CALCIUM ION, GLYCEROL, ... | | Authors: | Nishitani, Y, Maruyama, Y, Itoh, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2011-12-05 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recognition of heteropolysaccharide alginate by periplasmic solute-binding proteins of a bacterial ABC transporter
Biochemistry, 51, 2012
|
|
3VLU
 
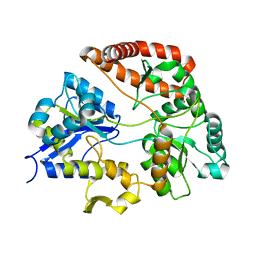 | | Crystal structure of Sphingomonas sp. A1 alginate-binding protein AlgQ1 in complex with saturated trimannuronate | | Descriptor: | AlgQ1, CALCIUM ION, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Nishitani, Y, Maruyama, Y, Itoh, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2011-12-05 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Recognition of heteropolysaccharide alginate by periplasmic solute-binding proteins of a bacterial ABC transporter
Biochemistry, 51, 2012
|
|
3VLV
 
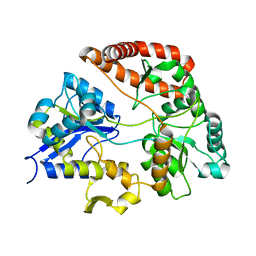 | | Crystal structure of Sphingomonas sp. A1 alginate-binding ptotein AlgQ1 in complex with unsaturated triguluronate | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid, AlgQ1, CALCIUM ION | | Authors: | Nishitani, Y, Maruyama, Y, Itoh, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2011-12-05 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Recognition of heteropolysaccharide alginate by periplasmic solute-binding proteins of a bacterial ABC transporter
Biochemistry, 51, 2012
|
|
3VXD
 
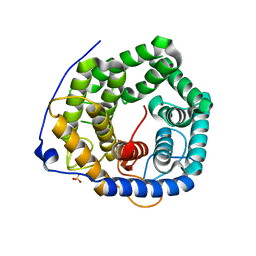 | | Crystal structure of unsaturated glucuronyl hydrolase mutant D115N from Streptcoccus agalactiae | | Descriptor: | Putative uncharacterized protein gbs1889, SULFATE ION | | Authors: | Nakamichi, Y, Maruyama, Y, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2012-09-11 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of unsaturated glucuronyl hydrolase mutant D115N from Streptcoccus agalactiae
To be Published
|
|
3WSC
 
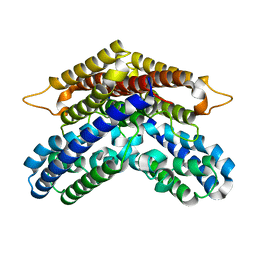 | |
3WUX
 
 | | Crystal structure of unsaturated glucuronyl hydrolase mutant D115N/K370S from Streptococcus agalactiae | | Descriptor: | 1,2-ETHANEDIOL, Unsaturated chondroitin disaccharide hydrolase | | Authors: | Nakamichi, Y, Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2014-05-08 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Crystal structure of unsaturated glucuronyl hydrolase mutant D115N/K370S from Streptococcus agalactiae
to be published
|
|
3WL3
 
 | | N,N'-diacetylchitobiose deacetylase from Pyrococcus horikoshii | | Descriptor: | GLYCEROL, PHOSPHATE ION, Putative uncharacterized protein PH0499, ... | | Authors: | Nakamura, T, Niiyama, M, Hashimoto, W, Uegaki, K. | | Deposit date: | 2013-11-07 | | Release date: | 2014-05-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Expression from engineered Escherichia coli chromosome and crystallographic study of archaeal N,N'-diacetylchitobiose deacetylase
Febs J., 281, 2014
|
|
3WIW
 
 | | Crystal structure of unsaturated glucuronyl hydrolase specific for heparin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Glycosyl hydrolase family 88 | | Authors: | Nakamichi, Y, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2013-09-26 | | Release date: | 2014-01-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of a bacterial unsaturated glucuronyl hydrolase with specificity for heparin.
J.Biol.Chem., 289, 2014
|
|
3WL4
 
 | | N,N'-diacetylchitobiose deacetylase (Se-derivative) from Pyrococcus furiosus | | Descriptor: | CADMIUM ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Nakamura, T, Niiyama, M, Hashimoto, W, Uegaki, K. | | Deposit date: | 2013-11-07 | | Release date: | 2014-05-07 | | Last modified: | 2014-08-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Expression from engineered Escherichia coli chromosome and crystallographic study of archaeal N,N'-diacetylchitobiose deacetylase
Febs J., 281, 2014
|
|
3VWO
 
 | |
