7VWT
 
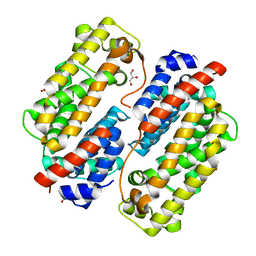 | | Carbazole Prenyl Transferase CqsB4 | | Descriptor: | 2-methyl-1-[(2R)-2-oxidanylpropyl]-9H-carbazole-3,4-dione, CqsB4, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Suemune, H, Nagata, R, Kuzuyama, T, Nagano, S. | | Deposit date: | 2021-11-11 | | Release date: | 2022-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural Basis for the Prenylation Reaction of Carbazole-Containing Natural Products Catalyzed by Squalene Synthase-Like Enzymes.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
3AJD
 
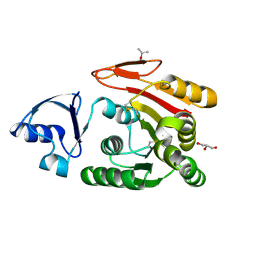 | | Crystal structure of ATRM4 | | Descriptor: | GLYCEROL, ISOPROPYL ALCOHOL, Putative methyltransferase MJ0026 | | Authors: | Hirano, M, Kuratani, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-06-01 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Crystal structure of Methanocaldococcus jannaschii Trm4 complexed with sinefungin.
J.Mol.Biol., 401, 2010
|
|
3VSN
 
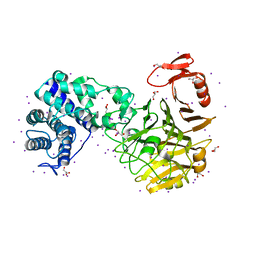 | | The crystal structure of novel chondroition lyase ODV-E66, baculovirus envelope protein | | Descriptor: | GLYCEROL, IODIDE ION, Occlusion-derived virus envelope protein E66 | | Authors: | Kawaguchi, Y, Sugiura, N, Kimata, K, Kimura, M, Kakuta, Y. | | Deposit date: | 2012-04-27 | | Release date: | 2013-05-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of novel chondroition lyase ODV-E66, baculovirus envelope protein
To be Published
|
|
3W1D
 
 | |
3W1C
 
 | |
3AZQ
 
 | |
3AZO
 
 | | Crystal structure of puromycin hydrolase | | Descriptor: | Aminopeptidase, SULFATE ION | | Authors: | Matoba, Y, Sugiyama, M. | | Deposit date: | 2011-05-27 | | Release date: | 2011-07-27 | | Last modified: | 2011-09-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence that puromycin hydrolase is a new type of aminopeptidase with a prolyl oligopeptidase family fold
Proteins, 79, 2011
|
|
3AZP
 
 | |
4MKC
 
 | | Crystal Structure of Anaplastic Lymphoma Kinase Complexed with LDK378 | | Descriptor: | 5-chloro-N~2~-[5-methyl-4-(piperidin-4-yl)-2-(propan-2-yloxy)phenyl]-N~4~-[2-(propan-2-ylsulfonyl)phenyl]pyrimidine-2,4-diamine, ALK tyrosine kinase receptor, GLYCEROL | | Authors: | Lee, C.C, Spraggon, G. | | Deposit date: | 2013-09-04 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The ALK Inhibitor Ceritinib Overcomes Crizotinib Resistance in Non-Small Cell Lung Cancer.
Cancer Discov, 4, 2014
|
|
4NNP
 
 | |
4NNO
 
 | |
2YY8
 
 | |
2YTZ
 
 | | Complex structure of Trm1 from Pyrococcus horikoshii with S-adenosyl-L-Homocystein in the orthorhombic crystal-lattice | | Descriptor: | N(2),N(2)-dimethylguanosine tRNA methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION | | Authors: | Ihsanawati, Shirouzu, M, Bessho, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2007-10-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structure of tRNA N(2),N(2)-Guanosine Dimethyltransferase Trm1 from Pyrococcus horikoshii
J.Mol.Biol., 383, 2008
|
|
3UMV
 
 | | Eukaryotic Class II CPD photolyase structure reveals a basis for improved UV-tolerance in plants | | Descriptor: | 1,2-ETHANEDIOL, Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Arvai, A.S, Hitomi, K, Getzoff, E.D, Tainer, J.A. | | Deposit date: | 2011-11-14 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.705 Å) | | Cite: | Eukaryotic Class II Cyclobutane Pyrimidine Dimer Photolyase Structure Reveals Basis for Improved Ultraviolet Tolerance in Plants.
J.Biol.Chem., 287, 2012
|
|
1MLD
 
 | | REFINED STRUCTURE OF MITOCHONDRIAL MALATE DEHYDROGENASE FROM PORCINE HEART AND THE CONSENSUS STRUCTURE FOR DICARBOXYLIC ACID OXIDOREDUCTASES | | Descriptor: | CITRIC ACID, MALATE DEHYDROGENASE | | Authors: | Gleason, W.B, Fu, Z, Birktoft, J.J, Banaszak, L.J. | | Deposit date: | 1994-01-24 | | Release date: | 1995-01-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Refined crystal structure of mitochondrial malate dehydrogenase from porcine heart and the consensus structure for dicarboxylic acid oxidoreductases.
Biochemistry, 33, 1994
|
|
2Z98
 
 | |
2Z9C
 
 | |
2Z9D
 
 | |
2Z9B
 
 | |
7WBM
 
 | | Crystal structure of Legionella pneumophila effector protein Lpg0081 | | Descriptor: | Lpg0081, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Lee, J, Kim, H, Oh, B.H. | | Deposit date: | 2021-12-17 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Reversible modification of mitochondrial ADP/ATP translocases by paired Legionella effector proteins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WBK
 
 | |
7XGO
 
 | | Human renin in complex with compound2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Renin, UNKNOWN LIGAND | | Authors: | Kashima, A. | | Deposit date: | 2022-04-05 | | Release date: | 2022-08-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of Novel 2-Carbamoyl Morpholine Derivatives as Highly Potent and Orally Active Direct Renin Inhibitors.
Acs Med.Chem.Lett., 13, 2022
|
|
7XGK
 
 | | Human renin in complex with compound1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Renin, UNKNOWN LIGAND | | Authors: | Kashima, A. | | Deposit date: | 2022-04-05 | | Release date: | 2022-08-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of Novel 2-Carbamoyl Morpholine Derivatives as Highly Potent and Orally Active Direct Renin Inhibitors.
Acs Med.Chem.Lett., 13, 2022
|
|
6JQA
 
 | |
3GFL
 
 | |
