5AEW
 
 | | Crystal structure of II9 variant of Biphenyl dioxygenase from Burkholderia xenovorans LB400 in complex with biphenyl | | Descriptor: | BIPHENYL, BIPHENYL DIOXYGENASE SUBUNIT ALPHA, BIPHENYL DIOXYGENASE SUBUNIT BETA, ... | | Authors: | Dhindwal, S, Gomez-Gil, L, Sylvestre, M, Eltis, L.D, Bolin, J.T, Kumar, P. | | Deposit date: | 2015-01-10 | | Release date: | 2016-04-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural Basis of the Enhanced Pollutant-Degrading Capabilities of an Engineered Biphenyl Dioxygenase.
J.Bacteriol., 198, 2016
|
|
4I14
 
 | | Crystal Structure of Mtb-ribA2 (Rv1415) | | Descriptor: | Riboflavin biosynthesis protein RibBA, SULFATE ION, ZINC ION | | Authors: | Singh, M, Kumar, P, Yadav, S, Gautam, R, Sharma, N, Karthikeyan, S. | | Deposit date: | 2012-11-20 | | Release date: | 2013-08-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure reveals the molecular mechanism of bifunctional 3,4-dihydroxy-2-butanone 4-phosphate synthase/GTP cyclohydrolase II (Rv1415) from Mycobacterium tuberculosis
Acta Crystallogr.,Sect.D, 69, 2013
|
|
9J7H
 
 | | Crystal structure of 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase (DAHP synthase) from Providencia alcalifaciens complexed with quinic acid | | Descriptor: | (1S,3R,4S,5R)-1,3,4,5-tetrahydroxycyclohexanecarboxylic acid, Phospho-2-dehydro-3-deoxyheptonate aldolase | | Authors: | Jangid, K, Mahto, J.K, Kumar, K.A, Kumar, P. | | Deposit date: | 2024-08-19 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural and biochemical analyses reveal quinic acid inhibits DAHP synthase a key player in shikimate pathway.
Arch.Biochem.Biophys., 763, 2025
|
|
9J7S
 
 | | Crystal structure of 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase (DAHP synthase) from Providencia alcalifaciens complexed with Phe | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHENYLALANINE, Phospho-2-dehydro-3-deoxyheptonate aldolase | | Authors: | Jangid, K, Mahto, J.K, Kumar, K.A, Kumar, P. | | Deposit date: | 2024-08-19 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and biochemical analyses reveal quinic acid inhibits DAHP synthase a key player in shikimate pathway.
Arch.Biochem.Biophys., 763, 2025
|
|
9J5B
 
 | |
9J1E
 
 | |
9J1V
 
 | |
9J1G
 
 | |
9J5C
 
 | |
9J59
 
 | |
9J58
 
 | |
9J57
 
 | |
9J5D
 
 | |
9J5A
 
 | |
5ZH8
 
 | | Crystal Structure of FmtA from Staphylococcus aureus at 2.58 A | | Descriptor: | GLYCEROL, Protein FmtA | | Authors: | Dalal, V, Kumar, P, Golemi-Kotra, D, Kumar, P. | | Deposit date: | 2018-03-12 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Repurposing an Ancient Protein Core Structure: Structural Studies on FmtA, a Novel Esterase of Staphylococcus aureus.
J.Mol.Biol., 431, 2019
|
|
9J19
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor minocycline | | Descriptor: | (4S,4AS,5AR,12AS)-4,7-BIS(DIMETHYLAMINO)-3,10,12,12A-TETRAHYDROXY-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2- CARBOXAMIDE, 3C-like proteinase nsp5 | | Authors: | Singh, A, Jangid, K, Dhaka, P, Tomar, S, Kumar, P. | | Deposit date: | 2024-08-04 | | Release date: | 2025-02-12 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Mechanistic Insights into the Main Protease (Mpro) Dimer Interface Destabilization Inhibitor: Unveiling New Therapeutic Avenues against SARS-CoV-2.
Biochemistry, 64, 2025
|
|
9IN1
 
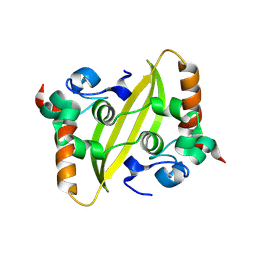 | |
6KM8
 
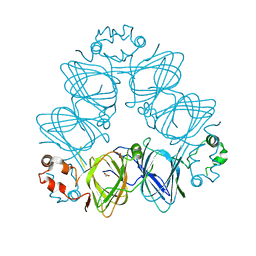 | | Crystal Structure of Momordica charantia 7S globulin | | Descriptor: | 7S globulin, ACETATE ION, COPPER (II) ION | | Authors: | Kesari, P, Pratap, S, Dhankhar, P, Dalal, V, Kumar, P. | | Deposit date: | 2019-07-31 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.099 Å) | | Cite: | Structural characterization and in-silico analysis of Momordica charantia 7S globulin for stability and ACE inhibition.
Sci Rep, 10, 2020
|
|
6LK2
 
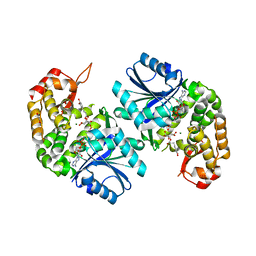 | | Crystal structure of Providencia alcalifaciens 3-dehydroquinate synthase (DHQS) in complex with Mg2+, NAD and chlorogenic acid | | Descriptor: | (1R,3R,4S,5R)-3-[3-[3,4-bis(oxidanyl)phenyl]propanoyloxy]-1,4,5-tris(oxidanyl)cyclohexane-1-carboxylic acid, 1,2-ETHANEDIOL, 3-dehydroquinate synthase, ... | | Authors: | Neetu, N, Katiki, M, Kumar, P. | | Deposit date: | 2019-12-17 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Structural and Biochemical Analyses Reveal that Chlorogenic Acid Inhibits the Shikimate Pathway.
J.Bacteriol., 202, 2020
|
|
6KMM
 
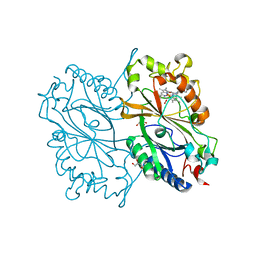 | | Crystal Structure of HEPES bound Dye Decolorizing peroxidase from Bacillus subtilis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Dhankhar, P, Dalal, V, Mahto, J.K, Kumar, P. | | Deposit date: | 2019-07-31 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Characterization of dye-decolorizing peroxidase from Bacillus subtilis.
Arch.Biochem.Biophys., 693, 2020
|
|
6KMN
 
 | | Crystal Structure of Dye Decolorizing peroxidase from Bacillus subtilis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Deferrochelatase/peroxidase EfeB, ... | | Authors: | Dhankhar, P, Dalal, V, Mahto, J.K, Kumar, P. | | Deposit date: | 2019-07-31 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Characterization of dye-decolorizing peroxidase from Bacillus subtilis.
Arch.Biochem.Biophys., 693, 2020
|
|
2GET
 
 | | Pantothenate kinase from Mycobacterium tuberculosis (MtPanK) in complex with a coenzyme A derivative, Form-I (LT) | | Descriptor: | GLYCEROL, Pantothenate kinase, [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-4-HYDROXY-3-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL (3R)-3-HYDROXY-4-{[3-({2-[(2-HYDROXYETHYL)DITHIO]ETHYL}AMINO)-3-OXOPROPYL]AMINO}-2,2-DIMETHYL-4-OXOBUTYL DIHYDROGEN DIPHOSPHATE | | Authors: | Das, S, Kumar, P, Bhor, V, Surolia, A, Vijayan, M. | | Deposit date: | 2006-03-20 | | Release date: | 2006-06-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Invariance and variability in bacterial PanK: a study based on the crystal structure of Mycobacterium tuberculosis PanK.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2GEU
 
 | | Pantothenate kinase from Mycobacterium tuberculosis (MtPanK) in complex with a coenzyme A derivative, Form-II (RT) | | Descriptor: | Pantothenate kinase, [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-4-HYDROXY-3-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL (3R)-3-HYDROXY-4-{[3-({2-[(2-HYDROXYETHYL)DITHIO]ETHYL}AMINO)-3-OXOPROPYL]AMINO}-2,2-DIMETHYL-4-OXOBUTYL DIHYDROGEN DIPHOSPHATE | | Authors: | Das, S, Kumar, P, Bhor, V, Surolia, A, Vijayan, M. | | Deposit date: | 2006-03-20 | | Release date: | 2006-06-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Invariance and variability in bacterial PanK: a study based on the crystal structure of Mycobacterium tuberculosis PanK.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2GEV
 
 | | Pantothenate kinase from Mycobacterium tuberculosis (MtPanK) in complex with a coenzyme A derivative, Form-II (LT) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Pantothenate kinase, ... | | Authors: | Das, S, Kumar, P, Bhor, V, Surolia, A, Vijayan, M. | | Deposit date: | 2006-03-20 | | Release date: | 2006-06-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Invariance and variability in bacterial PanK: a study based on the crystal structure of Mycobacterium tuberculosis PanK.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2GES
 
 | | Pantothenate kinase from Mycobacterium tuberculosis (MtPanK) in complex with a coenzyme A derivative, Form-I (RT) | | Descriptor: | Pantothenate kinase, [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-4-HYDROXY-3-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL (3R)-3-HYDROXY-4-{[3-({2-[(2-HYDROXYETHYL)DITHIO]ETHYL}AMINO)-3-OXOPROPYL]AMINO}-2,2-DIMETHYL-4-OXOBUTYL DIHYDROGEN DIPHOSPHATE | | Authors: | Das, S, Kumar, P, Bhor, V, Surolia, A, Vijayan, M. | | Deposit date: | 2006-03-20 | | Release date: | 2006-06-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Invariance and variability in bacterial PanK: a study based on the crystal structure of Mycobacterium tuberculosis PanK.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
