7KNX
 
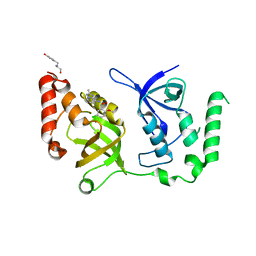 | | Crystal structure of SND1 in complex with C-26-A6 | | Descriptor: | 5-chloro-2-methoxy-N-(2-methyl[1,2,4]triazolo[1,5-a]pyridin-8-yl)benzene-1-sulfonamide, GLYCEROL, Staphylococcal nuclease domain-containing protein 1, ... | | Authors: | Kang, Y. | | Deposit date: | 2020-11-06 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Small-molecule inhibitors that disrupt the MTDH-SND1 complex suppress breast cancer progression and metastasis.
Nat Cancer, 3, 2022
|
|
7KNW
 
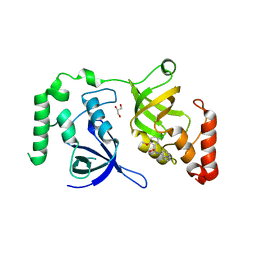 | | Crystal structure of SND1 in complex with C-26-A2 | | Descriptor: | 5-chloro-2-methoxy-N-([1,2,4]triazolo[1,5-a]pyridin-8-yl)benzene-1-sulfonamide, GLYCEROL, Staphylococcal nuclease domain-containing protein 1 | | Authors: | Kang, Y. | | Deposit date: | 2020-11-06 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Small-molecule inhibitors that disrupt the MTDH-SND1 complex suppress breast cancer progression and metastasis.
Nat Cancer, 3, 2022
|
|
3GW8
 
 | |
3GP5
 
 | |
3FTS
 
 | | Leukotriene A4 hydrolase in complex with resveratrol | | Descriptor: | ACETATE ION, IMIDAZOLE, Leukotriene A-4 hydrolase, ... | | Authors: | Davies, D.R. | | Deposit date: | 2009-01-13 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Discovery of leukotriene A4 hydrolase inhibitors using metabolomics biased fragment crystallography.
J.Med.Chem., 52, 2009
|
|
3FU6
 
 | |
3FUN
 
 | |
3FTW
 
 | |
3FUE
 
 | |
3FUD
 
 | |
3GP3
 
 | |
2LM4
 
 | | Solution NMR Structure of mitochondrial succinate dehydrogenase assembly factor 2 from Saccharomyces cerevisiae, Northeast Structural Genomics Consortium Target YT682A | | Descriptor: | Succinate dehydrogenase assembly factor 2, mitochondrial | | Authors: | Eletsky, A, Winge, D.R, Lee, H, Lee, D, Kohan, E, Hamilton, K, Acton, T.B, Xiao, R, Everett, J.K, Prestegard, J.H, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG), Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2011-11-22 | | Release date: | 2012-01-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of yeast succinate dehydrogenase flavinylation factor sdh5 reveals a putative sdh1 binding site.
Biochemistry, 51, 2012
|
|
3LNT
 
 | |
5Z9G
 
 | | Crystal structure of KAI2 | | Descriptor: | Probable esterase KAI2 | | Authors: | Kim, K.L, Cha, J.S, Soh, M.S, Cho, H.S. | | Deposit date: | 2018-02-03 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A missense allele of KARRIKIN-INSENSITIVE2 impairs ligand-binding and downstream signaling in Arabidopsis thaliana.
J. Exp. Bot., 69, 2018
|
|
4EN1
 
 | |
5Z9H
 
 | | Crystal structure of KAI2_ply2(A219V) | | Descriptor: | Probable esterase KAI2 | | Authors: | Kim, K.L, Cha, J.S, Soh, M.S, Cho, H.S. | | Deposit date: | 2018-02-03 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A missense allele of KARRIKIN-INSENSITIVE2 impairs ligand-binding and downstream signaling in Arabidopsis thaliana.
J. Exp. Bot., 69, 2018
|
|
4FJO
 
 | | Structure of the Rev1 CTD-Rev3/7-Pol kappa RIR complex | | Descriptor: | DNA polymerase kappa, DNA polymerase zeta catalytic subunit, DNA repair protein REV1, ... | | Authors: | Wojtaszek, J, Lee, C.-J, Zhou, P. | | Deposit date: | 2012-06-11 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.718 Å) | | Cite: | Structural basis of Rev1-mediated assembly of a quaternary vertebrate translesion polymerase complex consisting of Rev1, heterodimeric Pol zeta and Pol kappa
J.Biol.Chem., 287, 2012
|
|
3NNU
 
 | | Crystal structure of P38 alpha in complex with DP1376 | | Descriptor: | 2-{3-[(5E)-5-{[(2,3-dichlorophenyl)carbamoyl]imino}-3-thiophen-2-yl-2,5-dihydro-1H-pyrazol-1-yl]phenyl}acetamide, Mitogen-activated protein kinase 14 | | Authors: | Abendroth, J. | | Deposit date: | 2010-06-24 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Switch control pocket inhibitors of p38-MAP kinase. Durable type II inhibitors that do not require binding into the canonical ATP hinge region
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3NNX
 
 | | Crystal structure of phosphorylated P38 alpha in complex with DP802 | | Descriptor: | 2-[3-(3-tert-butyl-5-{[(2,3-dichlorophenyl)carbamoyl]imino}-2,5-dihydro-1H-pyrazol-1-yl)phenyl]acetamide, Mitogen-activated protein kinase 14 | | Authors: | Abendroth, J. | | Deposit date: | 2010-06-24 | | Release date: | 2010-09-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Switch control pocket inhibitors of p38-MAP kinase. Durable type II inhibitors that do not require binding into the canonical ATP hinge region
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3NNW
 
 | | Crystal structure of P38 alpha in complex with DP802 | | Descriptor: | 2-[3-(3-tert-butyl-5-{[(2,3-dichlorophenyl)carbamoyl]imino}-2,5-dihydro-1H-pyrazol-1-yl)phenyl]acetamide, Mitogen-activated protein kinase 14 | | Authors: | Abendroth, J. | | Deposit date: | 2010-06-24 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Switch control pocket inhibitors of p38-MAP kinase. Durable type II inhibitors that do not require binding into the canonical ATP hinge region
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3NNV
 
 | | Crystal structure of P38 alpha in complex with DP437 | | Descriptor: | 1-{3-tert-butyl-1-[4-(hydroxymethyl)phenyl]-1H-pyrazol-5-yl}-3-naphthalen-1-ylurea, Mitogen-activated protein kinase 14 | | Authors: | Abendroth, J. | | Deposit date: | 2010-06-24 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Switch control pocket inhibitors of p38-MAP kinase. Durable type II inhibitors that do not require binding into the canonical ATP hinge region
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3EZN
 
 | |
3FDZ
 
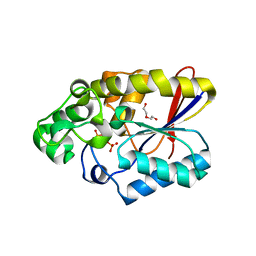 | | Crystal structure of phosphoglyceromutase from burkholderia pseudomallei 1710b with bound 2,3-diphosphoglyceric acid and 3-phosphoglyceric acid | | Descriptor: | (2R)-2,3-diphosphoglyceric acid, 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase, 3-PHOSPHOGLYCERIC ACID, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2008-11-26 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | An ensemble of structures of Burkholderia pseudomallei 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3FTY
 
 | |
3FU3
 
 | |
