8J6F
 
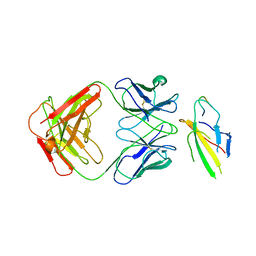 | | Cryo-EM structure of the Tocilizumab Fab/IL-6R complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Tocilizumab Fab, IL6R-D2 peptide, ... | | Authors: | She, J, Chen, L. | | Deposit date: | 2023-04-25 | | Release date: | 2024-03-27 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into IL-6 signaling inhibition by therapeutic antibodies.
Cell Rep, 43, 2024
|
|
8IOW
 
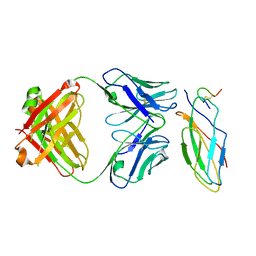 | | Cryo-EM structure of the sarilumab Fab/IL-6R complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Sarilumab Fab, Interleukin-6 receptor subunit alpha, ... | | Authors: | She, J, Chen, L, Wang, M.X. | | Deposit date: | 2023-03-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into IL-6 signaling inhibition by therapeutic antibodies.
Cell Rep, 43, 2024
|
|
6J66
 
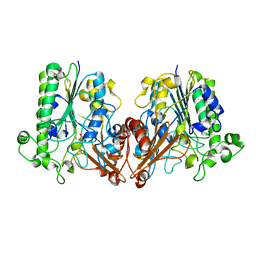 | | Chondroitin sulfate/dermatan sulfate endolytic 4-O-sulfatase | | Descriptor: | CALCIUM ION, Chondroitin sulfate/dermatan sulfate 4-O-endosulfatase protein | | Authors: | Gu, L, Li, F, Su, T, Wang, S. | | Deposit date: | 2019-01-14 | | Release date: | 2019-07-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Comparative Study of Two Chondroitin Sulfate/Dermatan Sulfate 4-O-Sulfatases With High Identity.
Front Microbiol, 10, 2019
|
|
7W8S
 
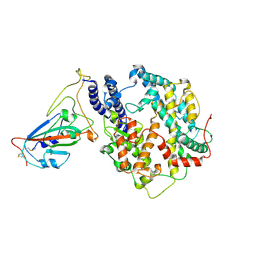 | | Structure of SARS-CoV-2 spike receptor-binding domain Y453F mutation complexed with American mink ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Su, C, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-12-08 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Molecular Basis of Mink ACE2 Binding to SARS-CoV-2 and Its Mink-Derived Variants.
J.Virol., 96, 2022
|
|
7WA1
 
 | |
7DWO
 
 | |
7DWN
 
 | | Crystal structure of Vibrio fischeri DarR in complex with DNA reveals the transcriptional activation mechanism of LTTR family members | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Predicted DNA-binding transcriptional regulator | | Authors: | Wang, W.W, Wu, H, He, J.H, Yu, F. | | Deposit date: | 2021-01-17 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Crystal structure details of Vibrio fischeri DarR and mutant DarR-M202I from LTTR family reveals their activation mechanism.
Int.J.Biol.Macromol., 183, 2021
|
|
8HI5
 
 | |
8HI6
 
 | |
5EAJ
 
 | | Crystal structure of DHFR in 0% Isopropanol | | Descriptor: | CALCIUM ION, CHLORIDE ION, Dihydrofolate reductase, ... | | Authors: | Cuneo, M.J, Agarwal, P.K. | | Deposit date: | 2015-10-16 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Modulating Enzyme Activity by Altering Protein Dynamics with Solvent.
Biochemistry, 57, 2018
|
|
8XLR
 
 | | Cryo-EM structure of Ca2+-bound TMEM16A in complex with Tamsulosin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Anoctamin-1, CALCIUM ION, ... | | Authors: | Li, H.L, Li, S.L, Li, S. | | Deposit date: | 2023-12-26 | | Release date: | 2024-12-18 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Tamsulosin ameliorates bone loss by inhibiting the release of Cl - through wedging into an allosteric site of TMEM16A.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
3UQP
 
 | | Crystal structure of Bace1 with its inhibitor | | Descriptor: | Beta-secretase 1, METHYL (2R)-1-[(6S,9S,12S,13S,17S,20S,23R)-9-(3-AMINO-3-OXOPROPYL)-12,23-DIBENZYL-13-HYDROXY-2,2,8,20,22-PENTAMETHYL-17-(2-METHYLPROPYL)-4,7,10,15,18,21,24-HEPTAOXO-6-(PROPAN-2-YL)-3-OXA-5,8,11,16,19,22-HEXAAZATETRACOSAN-24-YL]PYRROLIDINE-2-CARBOXYLATE, SULFATE ION | | Authors: | Chen, T.T, Chen, W.Y, Li, L, Xu, Y.C. | | Deposit date: | 2011-11-21 | | Release date: | 2012-11-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Cyanobacterial Peptides as a Prototype for the Design of Potent beta-Secretase Inhibitors and the Development of Selective Chemical Probes for Other Aspartic Proteases
J.Med.Chem., 55, 2012
|
|
4FGX
 
 | | Crystal structure of bace1 with novel inhibitor | | Descriptor: | Beta-secretase 1, DI(HYDROXYETHYL)ETHER, Inhibitor (2R,5S,8S,12S,13S,16S,19S,22S)-16-(3-amino-3-oxopropyl)-2,13-dibenzyl-12,22-dihydroxy-8-isobutyl-19-isopropyl-3,5,17-trimethyl-4,7,10,15,18,21-hexaoxo-3,6,9,14,17,20-hexaazatricosan-1-oic acid, ... | | Authors: | Chen, T.T, Chen, W.Y, Li, L, Xu, Y.C. | | Deposit date: | 2012-06-05 | | Release date: | 2013-01-16 | | Last modified: | 2021-09-15 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Cyanobacterial Peptides as a Prototype for the Design of Potent beta-Secretase Inhibitors and the Development of Selective Chemical Probes for Other Aspartic Proteases
J.Med.Chem., 55, 2012
|
|
8ZY2
 
 | | Sarbecovirus BANAL-20-52 Spike Trimer in a Locked Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Wang, J, Xiong, X. | | Deposit date: | 2024-06-16 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | SARS-related coronavirus S-protein structures reveal synergistic RBM interactions underpinning high-affinity human ACE2 binding.
Sci Adv, 11, 2025
|
|
8ZY9
 
 | | Ra9479 Bat ACE2 Dimer in Complex with Two BtKY72 Sarbecovirus Spike RBDs. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Wang, J, Xiong, X. | | Deposit date: | 2024-06-16 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | SARS-related coronavirus S-protein structures reveal synergistic RBM interactions underpinning high-affinity human ACE2 binding.
Sci Adv, 11, 2025
|
|
8ZY7
 
 | | Sarbecovirus HeB2013 Spike Trimer in a Locked Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, J, Xiong, X. | | Deposit date: | 2024-06-16 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | SARS-related coronavirus S-protein structures reveal synergistic RBM interactions underpinning high-affinity human ACE2 binding.
Sci Adv, 11, 2025
|
|
8ZY4
 
 | | Sarbecovirus YN2013 Spike Trimer in a Locked Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, ... | | Authors: | Wang, J, Xiong, X. | | Deposit date: | 2024-06-16 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | SARS-related coronavirus S-protein structures reveal synergistic RBM interactions underpinning high-affinity human ACE2 binding.
Sci Adv, 11, 2025
|
|
8ZY6
 
 | | Sarbecovirus GX2013 Spike Trimer in a Locked Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, J, Xiong, X. | | Deposit date: | 2024-06-16 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | SARS-related coronavirus S-protein structures reveal synergistic RBM interactions underpinning high-affinity human ACE2 binding.
Sci Adv, 11, 2025
|
|
8ZY3
 
 | | Sarbecovirus BANAL-20-236 Spike Trimer in a Locked Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, J, Xiong, X. | | Deposit date: | 2024-06-16 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | SARS-related coronavirus S-protein structures reveal synergistic RBM interactions underpinning high-affinity human ACE2 binding.
Sci Adv, 11, 2025
|
|
8ZY1
 
 | | Sarbecovirus BM48-31 Spike Trimer in a Locked Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Wang, J, Xiong, X. | | Deposit date: | 2024-06-16 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | SARS-related coronavirus S-protein structures reveal synergistic RBM interactions underpinning high-affinity human ACE2 binding.
Sci Adv, 11, 2025
|
|
8ZYA
 
 | | Ra9479 Bat ACE2 Bound to BtkY72 Sarbecovirus Spike RBD (Focused Refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Wang, J, Xiong, X. | | Deposit date: | 2024-06-16 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | SARS-related coronavirus S-protein structures reveal synergistic RBM interactions underpinning high-affinity human ACE2 binding.
Sci Adv, 11, 2025
|
|
8ZY5
 
 | | Sarbecovirus RmYN02 Spike Trimer in a Locked Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, J, Xiong, X. | | Deposit date: | 2024-06-16 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | SARS-related coronavirus S-protein structures reveal synergistic RBM interactions underpinning high-affinity human ACE2 binding.
Sci Adv, 11, 2025
|
|
8ZY0
 
 | | Sarbecovirus BtKY72 Spike Trimer in a Locked Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Wang, J, Xiong, X. | | Deposit date: | 2024-06-16 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | SARS-related coronavirus S-protein structures reveal synergistic RBM interactions underpinning high-affinity human ACE2 binding.
Sci Adv, 11, 2025
|
|
7E9O
 
 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with 35B5 Fab(3 up RBDs, state2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, Light chain of 35B5 Fab, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-03-04 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
7E9Q
 
 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with 35B5 Fab(1 out RBD, state3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, Light chain of 35B5 Fab, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-03-04 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
