6UD0
 
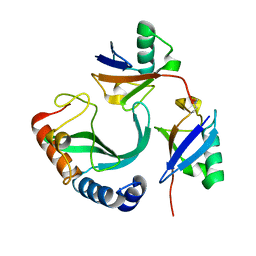 | | Solution-state NMR structural ensemble of human Tsg101 UEV in complex with K63-linked diubiquitin | | Descriptor: | Tumor susceptibility gene 101 protein, Ubiquitin | | Authors: | Strickland, M, Watanabe, S, Bonn, S.M, Camara, C.M, Fushman, D, Carter, C.A, Tjandra, N. | | Deposit date: | 2019-09-18 | | Release date: | 2021-03-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Tsg101/ESCRT-I Recruitment Regulated by the Dual Binding Modes of K63-Linked Diubiquitin
Structure, 2021
|
|
5JOE
 
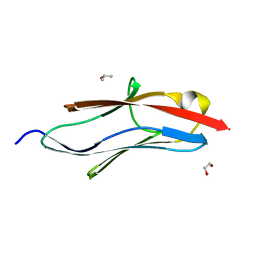 | | Crystal structure of I81 from titin | | Descriptor: | 1,2-ETHANEDIOL, ISOPROPYL ALCOHOL, Titin | | Authors: | Fleming, J, Zhou, T, Bogomolovas, J, Labeit, S, Mayans, O. | | Deposit date: | 2016-05-02 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CARP interacts with titin at a unique helical N2A sequence and at the domain Ig81 to form a structured complex.
Febs Lett., 590, 2016
|
|
7X9U
 
 | | Type-II KH motif of human mitochondrial RbfA | | Descriptor: | Putative ribosome-binding factor A, mitochondrial | | Authors: | Kuwasako, K, Suzuki, S, Furue, M, Takizawa, M, Takahashi, M, Tsuda, K, Nagata, T, Watanabe, S, Tanaka, A, Kobayashi, N, Kigawa, T, Guntert, P, Shirouzu, M, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2022-03-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1 H, 13 C, and 15 N resonance assignments and solution structures of the KH domain of human ribosome binding factor A, mtRbfA, involved in mitochondrial ribosome biogenesis.
Biomol.Nmr Assign., 16, 2022
|
|
5YXY
 
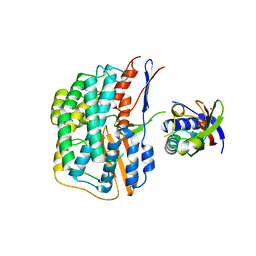 | | Crystal structure of the HyhL-HypA complex (form I) | | Descriptor: | Cytosolic NiFe-hydrogenase, alpha subunit, Probable hydrogenase nickel incorporation protein HypA, ... | | Authors: | Kwon, S, Watanabe, S, Nishitani, Y, Miki, K. | | Deposit date: | 2017-12-07 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.299 Å) | | Cite: | Crystal structures of a [NiFe] hydrogenase large subunit HyhL in an immature state in complex with a Ni chaperone HypA.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5YY0
 
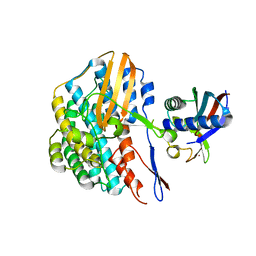 | | Crystal structure of the HyhL-HypA complex (form II) | | Descriptor: | Cytosolic NiFe-hydrogenase, alpha subunit, Probable hydrogenase nickel incorporation protein HypA, ... | | Authors: | Kwon, S, Watanabe, S, Nishitani, Y, Miki, K. | | Deposit date: | 2017-12-07 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.243 Å) | | Cite: | Crystal structures of a [NiFe] hydrogenase large subunit HyhL in an immature state in complex with a Ni chaperone HypA.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7Y9P
 
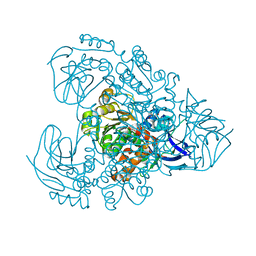 | | Xylitol dehydrogenase S96C/S99C/Y102C mutant(thermostabilized form) from Pichia stipitis | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SULFATE ION, ... | | Authors: | Yoshiwara, K, Watanabe, Y, Watanabe, S. | | Deposit date: | 2022-06-25 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular evolutionary insight of structural zinc atom in yeast xylitol dehydrogenases and its application in bioethanol production by lignocellulosic biomass.
Sci Rep, 13, 2023
|
|
6J7C
 
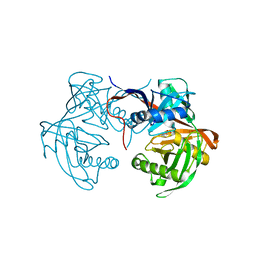 | | Crystal structure of proline racemase-like protein from Thermococcus litoralis in complex with proline | | Descriptor: | PROLINE, Proline racemase | | Authors: | Watanabe, Y, Watanabe, S, Itoh, Y, Watanabe, Y. | | Deposit date: | 2019-01-17 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of substrate-bound bifunctional proline racemase/hydroxyproline epimerase from a hyperthermophilic archaeon.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
7E7S
 
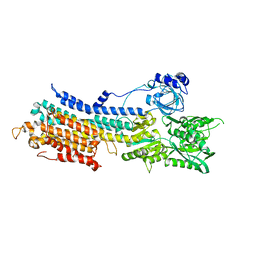 | | WT transporter state1 | | Descriptor: | CALCIUM ION, Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | | Authors: | Zhang, Y, Watanabe, S, Tsutsumi, A, Inaba, K. | | Deposit date: | 2021-02-27 | | Release date: | 2021-09-08 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM analysis provides new mechanistic insight into ATP binding to Ca 2+ -ATPase SERCA2b.
Embo J., 40, 2021
|
|
6JNJ
 
 | |
5U9D
 
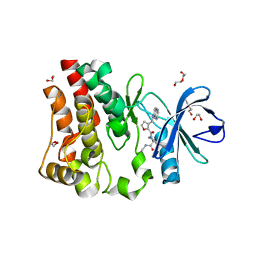 | | Discovery of a potent BTK inhibitor with a novel binding mode using parallel selections with a DNA-encoded chemical library | | Descriptor: | (R)-N-methyl-2-(3-((quinoxalin-6-ylamino)methyl)furan-2-carbonyl)-2,3,4,9-tetrahydro-1H-pyrido[3,4-b]indole-3-carboxamide, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Cuozzo, J.W, Centrella, P.A, Gikunju, D, Habeshian, S, Hupp, C.D, Keefe, A.D, Sigel, E, Soutter, H.H, Thomson, H.A, Zhang, Y, Clark, M.A. | | Deposit date: | 2016-12-16 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Discovery of a Potent BTK Inhibitor with a Novel Binding Mode by Using Parallel Selections with a DNA-Encoded Chemical Library.
Chembiochem, 18, 2017
|
|
7W7V
 
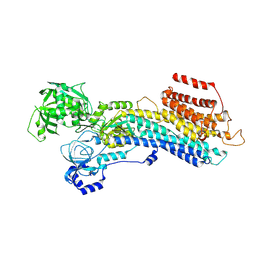 | | 'late' E2P of SERCA2b | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | | Authors: | Zhang, Y, Watanabe, S, Tsutsumi, A, Inaba, K. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Multiple sub-state structures of SERCA2b reveal conformational overlap at transition steps during the catalytic cycle.
Cell Rep, 41, 2022
|
|
7W7U
 
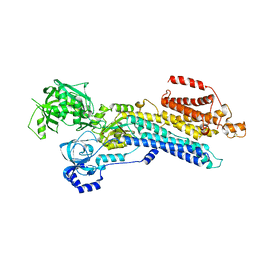 | | The 'Ca2+-unbound' BeF3- of SERCA2b | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | | Authors: | Zhang, Y, Watanabe, S, Tsutsumi, A, Inaba, K. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-14 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Multiple sub-state structures of SERCA2b reveal conformational overlap at transition steps during the catalytic cycle.
Cell Rep, 41, 2022
|
|
7W7W
 
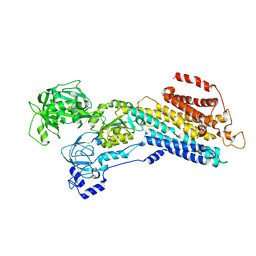 | | E2 Pi of SERCA2b | | Descriptor: | MAGNESIUM ION, Sarcoplasmic/endoplasmic reticulum calcium ATPase 2, TETRAFLUOROALUMINATE ION | | Authors: | Zhang, Y, Watanabe, S, Tsutsumi, A, Inaba, K. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-14 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Multiple sub-state structures of SERCA2b reveal conformational overlap at transition steps during the catalytic cycle.
Cell Rep, 41, 2022
|
|
7W7T
 
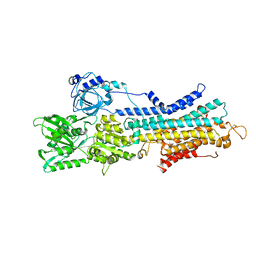 | | The E1-BeF3- 2Ca2+ of SERCA2b | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Zhang, Y, Watanabe, S, Tsutsumi, A, Inaba, K. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-14 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Multiple sub-state structures of SERCA2b reveal conformational overlap at transition steps during the catalytic cycle.
Cell Rep, 41, 2022
|
|
6JNK
 
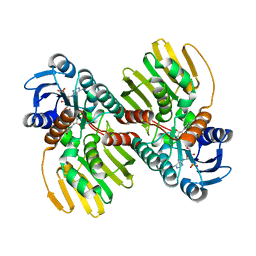 | |
5B25
 
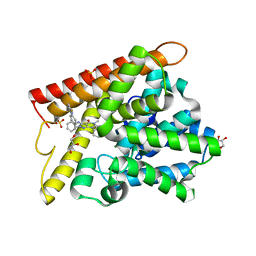 | | Crystal structure of human PDE1B with inhibitor 3 | | Descriptor: | (11R,15S)-4-{[4-(6-fluoropyridin-2-yl)phenyl]methyl}-8-methyl-5-(phenylamino)-1,3,4,8,10-pentaazatetracyclo[7.6.0.02,6.011,15]pentadeca-2,5,9-trien-7-one, Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B, GLYCEROL, ... | | Authors: | Ida, K, Lane, W, Snell, G, Sogabe, S. | | Deposit date: | 2016-01-07 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Potent and Selective Inhibitors of Phosphodiesterase 1 for the Treatment of Cognitive Impairment Associated with Neurodegenerative and Neuropsychiatric Diseases
J.Med.Chem., 59, 2016
|
|
6L06
 
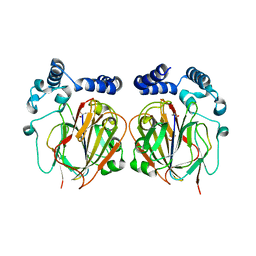 | |
6L07
 
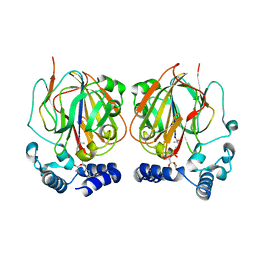 | |
1CRY
 
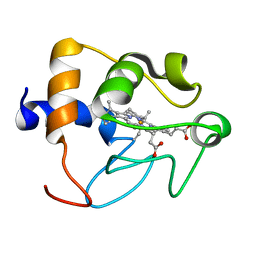 | |
2M1M
 
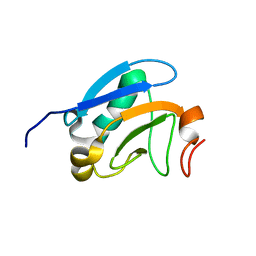 | | Solution structure of the PsIAA4 oligomerization domain reveals interaction modes for transcription factors in early auxin response | | Descriptor: | Auxin-induced protein IAA4 | | Authors: | Kovermann, M, Dinesh, D.C, Gopalswamy, M, Abel, S, Balbach, J. | | Deposit date: | 2012-12-03 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PsIAA4 oligomerization domain reveals interaction modes for transcription factors in early auxin response.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4JNW
 
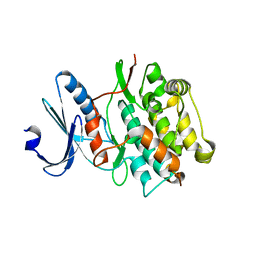 | | Bacterially expressed Titin Kinase | | Descriptor: | GLYCEROL, Titin | | Authors: | Bogomolovas, J, Labeit, S, Mayans, O. | | Deposit date: | 2013-03-16 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Titin kinase is an inactive pseudokinase scaffold that supports MuRF1 recruitment to the sarcomeric M-line.
Open Biol, 4, 2014
|
|
3VX3
 
 | | Crystal structure of [NiFe] hydrogenase maturation protein HypB from Thermococcus kodakarensis KOD1 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ATPase involved in chromosome partitioning, ... | | Authors: | Sasaki, D, Watanabe, S, Miki, K. | | Deposit date: | 2012-09-09 | | Release date: | 2013-02-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification and Structure of a Novel Archaeal HypB for [NiFe] Hydrogenase Maturation
J.Mol.Biol., 425, 2013
|
|
3WJQ
 
 | | Crystal structure of the HypE CN form | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, BENZAMIDINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tominaga, T, Watanabe, S, Miki, K. | | Deposit date: | 2013-10-14 | | Release date: | 2013-12-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.645 Å) | | Cite: | Crystal structures of the carbamoylated and cyanated forms of HypE for [NiFe] hydrogenase maturation
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3WJP
 
 | | Crystal structure of the HypE CA form | | Descriptor: | BENZAMIDINE, GLYCEROL, Hydrogenase expression/formation protein HypE, ... | | Authors: | Tominaga, T, Watanabe, S, Miki, K. | | Deposit date: | 2013-10-14 | | Release date: | 2013-12-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.533 Å) | | Cite: | Crystal structures of the carbamoylated and cyanated forms of HypE for [NiFe] hydrogenase maturation
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3WJR
 
 | | crystal structure of HypE in complex with a nucleotide | | Descriptor: | ADENOSINE MONOPHOSPHATE, BENZAMIDINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tominaga, T, Watanabe, S, Miki, K. | | Deposit date: | 2013-10-14 | | Release date: | 2013-12-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.864 Å) | | Cite: | Crystal structures of the carbamoylated and cyanated forms of HypE for [NiFe] hydrogenase maturation
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
