1TCO
 
 | | TERNARY COMPLEX OF A CALCINEURIN A FRAGMENT, CALCINEURIN B, FKBP12 AND THE IMMUNOSUPPRESSANT DRUG FK506 (TACROLIMUS) | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, CALCIUM ION, FE (III) ION, ... | | Authors: | Griffith, J.P, Kim, J.L, Kim, E.E, Sintchak, M.D, Thomson, J.A, Fitzgibbon, M.J, Fleming, M.A, Caron, P.R, Hsiao, K, Navia, M.A. | | Deposit date: | 1996-08-21 | | Release date: | 1997-02-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structure of calcineurin inhibited by the immunophilin-immunosuppressant FKBP12-FK506 complex.
Cell(Cambridge,Mass.), 82, 1995
|
|
1T15
 
 | | Crystal Structure of the Brca1 BRCT Domains in Complex with the Phosphorylated Interacting Region from Bach1 Helicase | | Descriptor: | BRCA1 interacting protein C-terminal helicase 1, Breast cancer type 1 susceptibility protein | | Authors: | Clapperton, J.A, Manke, I.A, Lowery, D.M, Ho, T, Haire, L.F, Yaffe, M.B, Smerdon, S.J. | | Deposit date: | 2004-04-15 | | Release date: | 2004-05-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and mechanism of BRCA1 BRCT domain recognition of phosphorylated BACH1 with implications for cancer
Nat.Struct.Mol.Biol., 11, 2004
|
|
1SSZ
 
 | | Conformational Mapping of Mini-B: An N-terminal/C-terminal Construct of Surfactant Protein B Using 13C-Enhanced Fourier Transform Infrared (FTIR) Spectroscopy | | Descriptor: | Pulmonary surfactant-associated protein B | | Authors: | Waring, A.J, Walther, F.J, Gordon, L.M, Hernandez-Juviel, J.M, Hong, T, Sherman, M.A, Alonso, C, Alig, T, Braun, A, Bacon, D, Zasadzinski, J.A. | | Deposit date: | 2004-03-24 | | Release date: | 2004-06-15 | | Last modified: | 2019-04-24 | | Method: | INFRARED SPECTROSCOPY | | Cite: | The role of charged amphipathic helices in the structure and function of surfactant protein B.
J.Pept.Res., 66, 2005
|
|
1TEU
 
 | | Effect of Shuttle Location and pH Environment on H+ Transfer in Human Carbonic Anhydrase II | | Descriptor: | Carbonic anhydrase II, ZINC ION | | Authors: | Fisher, Z, Hernandez Prada, J.A, Tu, C.K, Duda, D, Yoshioka, C, An, H, Govindasamy, L, Silverman, D.N, McKenna, R. | | Deposit date: | 2004-05-25 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Kinetic Characterization of Active-Site Histidine as a Proton Shuttle in Catalysis by Human Carbonic Anhydrase II
Biochemistry, 44, 2005
|
|
1SZU
 
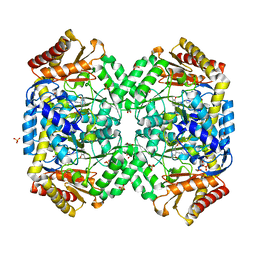 | | The structure of gamma-aminobutyrate aminotransferase mutant: V241A | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-aminobutyrate aminotransferase, ... | | Authors: | Liu, W, Peterson, P.E, Langston, J.A, Jin, X, Zhou, X, Fisher, A.J, Toney, M.D. | | Deposit date: | 2004-04-06 | | Release date: | 2005-03-01 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Kinetic and Crystallographic Analysis of Active Site Mutants of Escherichia coligamma-Aminobutyrate Aminotransferase.
Biochemistry, 44, 2005
|
|
1TKK
 
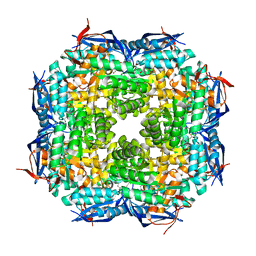 | | The Structure of a Substrate-Liganded Complex of the L-Ala-D/L-Glu Epimerase from Bacillus subtilis | | Descriptor: | ALANINE, GLUTAMIC ACID, MAGNESIUM ION, ... | | Authors: | Klenchin, V.A, Schmidt, D.M, Gerlt, J.A, Rayment, I. | | Deposit date: | 2004-06-08 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Evolution of Enzymatic Activities in the Enolase Superfamily: Structure of a Substrate-Liganded Complex of the l-Ala-d/l-Glu Epimerase from Bacillus subtilis(,).
Biochemistry, 43, 2004
|
|
7BHP
 
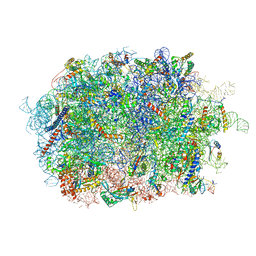 | | Cryo-EM structure of the human Ebp1 - 80S ribosome | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Desogus, J, Bhaskar, V, Chao, J.A. | | Deposit date: | 2021-01-11 | | Release date: | 2021-02-03 | | Last modified: | 2021-03-31 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Dynamic association of human Ebp1 with the ribosome.
Rna, 27, 2021
|
|
7BAY
 
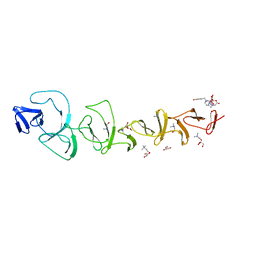 | |
7E6V
 
 | | The crystal structure of foot-and-mouth disease virus(FMDV) 2C protein 97-318aa | | Descriptor: | ACETATE ION, Protein 2C | | Authors: | Zhang, C, Wojdyla, J.A, Qin, B, Wang, M, Gao, X, Cui, S. | | Deposit date: | 2021-02-24 | | Release date: | 2022-06-29 | | Last modified: | 2022-07-20 | | Method: | X-RAY DIFFRACTION (1.832 Å) | | Cite: | An anti-picornaviral strategy based on the crystal structure of foot-and-mouth disease virus 2C protein.
Cell Rep, 40, 2022
|
|
7BKW
 
 | | Endothiapepsin structure obtained at 100K with fragment BTB09871 bound | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Engilberge, S, Huang, C.-Y, Smith, K.M.L, Eris, D, Marsh, M, Wang, M, Wojdyla, J.A. | | Deposit date: | 2021-01-17 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Endothiapepsin structure obtained at 100K with fragment BTB09871 bound
To Be Published
|
|
5K4B
 
 | | Structure of eukaryotic translation initiation factor 3 subunit D (eIF3d) cap binding domain from Nasonia vitripennis, Crystal form 1 | | Descriptor: | CHLORIDE ION, Eukaryotic translation initiation factor 3 subunit D | | Authors: | Kranzusch, P.J, Lee, A.S.Y, Doudna, J.A, Cate, J.H.D. | | Deposit date: | 2016-05-20 | | Release date: | 2016-07-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation.
Nature, 536, 2016
|
|
5K92
 
 | | Crystal Structure of an apo Tris-thiolate Binding Site in a de novo Three Stranded Coiled Coil Peptide | | Descriptor: | Apo-(CSL16C)3, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ruckthong, L, Zastrow, M.L, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2016-05-31 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | A Crystallographic Examination of Predisposition versus Preorganization in de Novo Designed Metalloproteins.
J.Am.Chem.Soc., 138, 2016
|
|
5JXP
 
 | | Crystal structure of Porphyromonas endodontalis DPP11 in alternate conformation | | Descriptor: | Asp/Glu-specific dipeptidyl-peptidase, CALCIUM ION, CHLORIDE ION | | Authors: | Bezerra, G.A, Cornaciu, I, Hoffmann, G, Djinovic-Carugo, K, Marquez, J.A. | | Deposit date: | 2016-05-13 | | Release date: | 2017-06-14 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Bacterial protease uses distinct thermodynamic signatures for substrate recognition.
Sci Rep, 7, 2017
|
|
5K4D
 
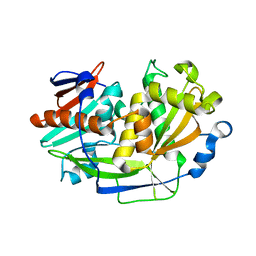 | | Structure of eukaryotic translation initiation factor 3 subunit D (eIF3d) cap binding domain from Nasonia vitripennis, Crystal form 3 | | Descriptor: | Eukaryotic translation initiation factor 3 subunit D | | Authors: | Kranzusch, P.J, Lee, A.S.Y, Doudna, J.A, Cate, J.H.D. | | Deposit date: | 2016-05-20 | | Release date: | 2016-07-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation.
Nature, 536, 2016
|
|
5KEE
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS L25K/I92F at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Skerritt, L.A, Caro, J.A, Heroux, A, Schlessman, J.L, Garcia-Moreno E, B. | | Deposit date: | 2016-06-09 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS L25K/I92F at cryogenic temperature
To be Published
|
|
5KCE
 
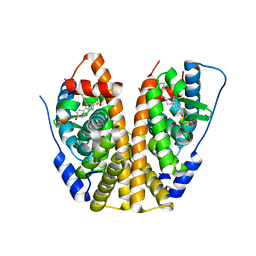 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with an N-methyl, 2-chlorobenzyl OBHS-N derivative | | Descriptor: | (1S,2R,4S)-N-(2-chlorophenyl)-5,6-bis(4-hydroxyphenyl)-N-methyl-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Dharmarajan, V, Goswami, D, Kastrati, I, Novick, S, Nowak, J, Zhou, H.B, Boonmuen, N, Zhao, Y, Min, J, Frasor, J, Katzenellenbogen, B.S, Griffin, P.R, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-06-06 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Full antagonism of the estrogen receptor without a prototypical ligand side chain.
Nat. Chem. Biol., 13, 2017
|
|
5JXZ
 
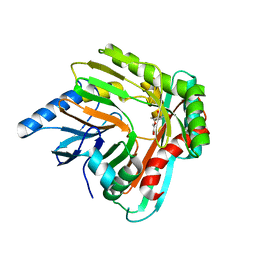 | | A low magnesium structure of the isochorismate synthase, EntC | | Descriptor: | (3R,4R)-3-[(1-carboxyethenyl)oxy]-4-hydroxycyclohexa-1,5-diene-1-carboxylic acid, (5S,6S)-5-[(1-carboxyethenyl)oxy]-6-hydroxycyclohexa-1,3-diene-1-carboxylic acid, Isochorismate synthase EntC, ... | | Authors: | Meneely, K.M, Sundlov, J.A, Gulick, A.M, Lamb, A.L. | | Deposit date: | 2016-05-13 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | An Open and Shut Case: The Interaction of Magnesium with MST Enzymes.
J.Am.Chem.Soc., 138, 2016
|
|
5KB0
 
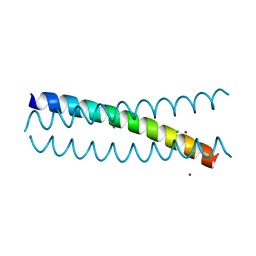 | | Crystal Structure of a Tris-thiolate Pb(II) Complex in a de Novo Three-stranded Coiled Coil Peptide | | Descriptor: | CHLORIDE ION, LEAD (II) ION, Pb(II)Zn(II)(GRAND Coil Ser-L16CL30H)3+, ... | | Authors: | Ruckthong, L, Zastrow, M.L, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2016-06-02 | | Release date: | 2016-08-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | A Crystallographic Examination of Predisposition versus Preorganization in de Novo Designed Metalloproteins.
J.Am.Chem.Soc., 138, 2016
|
|
5KCU
 
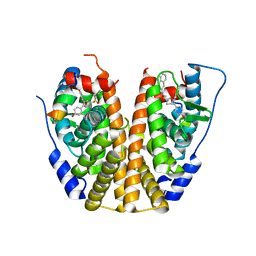 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with an N-ethyl, alpha-naphthyl OBHS-N derivative | | Descriptor: | (1S,2R,4S)-N-ethyl-5,6-bis(4-hydroxyphenyl)-N-(naphthalen-2-yl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor, NCOA2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Dharmarajan, V, Goswami, D, Kastrati, I, Novick, S, Nowak, J, Zhou, H.B, Boonmuen, N, Zhao, Y, Min, J, Frasor, J, Katzenellenbogen, B.S, Griffin, P.R, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-06-07 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Full antagonism of the estrogen receptor without a prototypical ligand side chain.
Nat. Chem. Biol., 13, 2017
|
|
5KCD
 
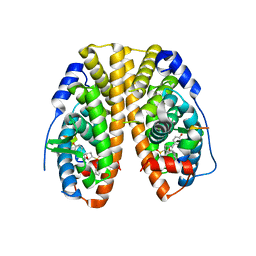 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with an N-methyl Substituted OBHS-N derivative | | Descriptor: | (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-N-methyl-N-phenyl-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor, NCOA2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Dharmarajan, V, Goswami, D, Kastrati, I, Novick, S, Nowak, J, Zhou, H.B, Boonmuen, N, Zhao, Y, Min, J, Frasor, J, Katzenellenbogen, B.S, Griffin, P.R, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-06-06 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Full antagonism of the estrogen receptor without a prototypical ligand side chain.
Nat. Chem. Biol., 13, 2017
|
|
5KCF
 
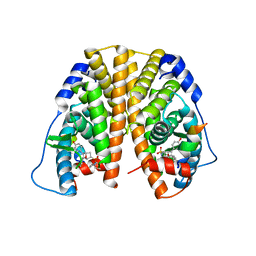 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with an N-ethyl, 4-methoxybenzyl OBHS-N derivative | | Descriptor: | (1R,2S,4R)-N-ethyl-5,6-bis(4-hydroxyphenyl)-N-(4-methoxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, (1S,2R,4S)-N-ethyl-5,6-bis(4-hydroxyphenyl)-N-(4-methoxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor, ... | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Dharmarajan, V, Goswami, D, Kastrati, I, Novick, S, Nowak, J, Zhou, H.B, Boonmuen, N, Zhao, Y, Min, J, Frasor, J, Katzenellenbogen, B.S, Griffin, P.R, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-06-06 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Full antagonism of the estrogen receptor without a prototypical ligand side chain.
Nat. Chem. Biol., 13, 2017
|
|
5KCT
 
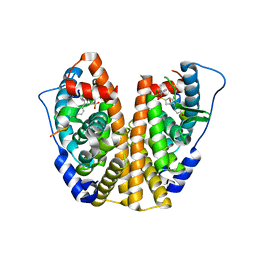 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with an N-ethyl, 4-chlorobenzyl OBHS-N derivative | | Descriptor: | (1R,2S,4R)-N-(4-chlorophenyl)-N-ethyl-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, (1S,2R,4S)-N-(4-chlorophenyl)-N-ethyl-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor, ... | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Dharmarajan, V, Goswami, D, Kastrati, I, Novick, S, Nowak, J, Zhou, H.B, Boonmuen, N, Zhao, Y, Min, J, Frasor, J, Katzenellenbogen, B.S, Griffin, P.R, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-06-07 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Full antagonism of the estrogen receptor without a prototypical ligand side chain.
Nat. Chem. Biol., 13, 2017
|
|
5KRL
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the A-CD ring estrogen, (1S,7aS)-5-(2-chloro-4-hydroxyphenyl)-7a-methyl-2,3,3a,4,7,7a-hexahydro-1H-inden-1-ol | | Descriptor: | (1~{S},3~{a}~{R},7~{a}~{S})-5-(2-chloranyl-4-oxidanyl-phenyl)-2,3,3~{a},4,7,7~{a}-hexahydro-1~{H}-inden-1-ol, Estrogen receptor, NCOA2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-07-07 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5KO0
 
 | | Human Islet Amyloid Polypeptide Segment 15-FLVHSSNNFGA-25 Determined by MicroED | | Descriptor: | THIOCYANATE ION, hIAPP(15-25)WT | | Authors: | Krotee, P.A.L, Rodriguez, J.A, Sawaya, M.R, Cascio, D, Shi, D, Nannenga, B.L, Hattne, J, Reyes, F.E, Gonen, T, Eisenberg, D.S. | | Deposit date: | 2016-06-28 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.4 Å) | | Cite: | Atomic structures of fibrillar segments of hIAPP suggest tightly mated beta-sheets are important for cytotoxicity.
Elife, 6, 2017
|
|
5KRM
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the A-CD ring estrogen, (1S,7aS)-5-(2,5-difluoro-4-hydroxyphenyl)-7a-methyl-2,3,3a,4,7,7a-hexahydro-1H-inden-1-ol | | Descriptor: | (1~{S},3~{a}~{R},7~{a}~{S})-5-[2,5-bis(fluoranyl)-4-oxidanyl-phenyl]-7~{a}-methyl-1,2,3,3~{a},4,7-hexahydroinden-1-ol, Estrogen receptor, NCOA2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-07-07 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
