7KFY
 
 | | Structural basis for a germline-biased antibody response to SARS-CoV-2 (RBD:C1A-F10 Fab) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, heavy chain of human antibody C1A-F10 Fab, ... | | Authors: | Pan, J, Abraham, J, Clark, L, Clark, S. | | Deposit date: | 2020-10-15 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.157 Å) | | Cite: | Molecular basis for a germline-biased neutralizing antibody response to SARS-CoV-2.
Biorxiv, 2020
|
|
6BBA
 
 | |
7KFW
 
 | | Structural basis for a germline-biased antibody response to SARS-CoV-2 (RBD:C1A-B3 Fab) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, heavy chain of antibody C1A-B3 Fab, ... | | Authors: | Pan, J, Abraham, J, Clark, L, Clark, S. | | Deposit date: | 2020-10-15 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.792 Å) | | Cite: | Molecular basis for a germline-biased neutralizing antibody response to SARS-CoV-2.
Biorxiv, 2020
|
|
5C9I
 
 | |
4KT1
 
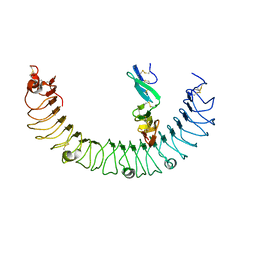 | | Complex of R-spondin 1 with LGR4 extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Leucine-rich repeat-containing G-protein coupled receptor 4, ... | | Authors: | Wang, X.Q, Wang, D.L. | | Deposit date: | 2013-05-19 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Structural basis for R-spondin recognition by LGR4/5/6 receptors
Genes Dev., 27, 2013
|
|
2WD4
 
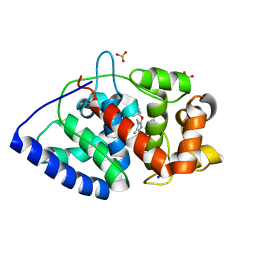 | | Ascorbate Peroxidase as a heme oxygenase: w41A variant product with t-butyl peroxide | | Descriptor: | 3-[2-[[3-(2-CARBOXYETHYL)-5-[[3-ETHENYL-4-METHYL-5-[(2-METHYLPROPAN-2-YL)OXY]-1H-PYRROL-2-YL]METHYL]-4-METHYL-1H-PYRROL -2-YL]METHYL]-5-[(Z)-(4-ETHENYL-3-METHYL-5-OXO-PYRROL-2-YLIDENE)METHYL]-4-METHYL-1H-PYRROL-3-YL]PROPANOIC ACID, ASCORBATE PEROXIDASE, FE (III) ION, ... | | Authors: | Badyal, S.K, Metcalfe, C.L, Gumiero, A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2009-03-19 | | Release date: | 2009-04-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Evidence for Heme Oxygenase Activity in a Heme Peroxidase.
Biochemistry, 48, 2009
|
|
5AVA
 
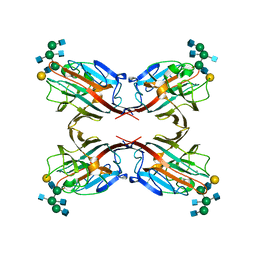 | |
5B1X
 
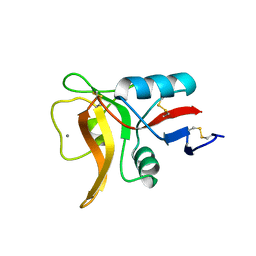 | |
7KFX
 
 | | Structural basis for a germline-biased antibody response to SARS-CoV-2 (RBD:C1A-C2 Fab) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, heavy chain of human antibody C1A-C2 Fab, ... | | Authors: | Pan, J, Abraham, J, Clark, L, Clark, S. | | Deposit date: | 2020-10-15 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.226 Å) | | Cite: | Molecular basis for a germline-biased neutralizing antibody response to SARS-CoV-2.
Biorxiv, 2020
|
|
5C8F
 
 | |
2FGE
 
 | |
5C8A
 
 | |
5C8E
 
 | |
1A19
 
 | | BARSTAR (FREE), C82A MUTANT | | Descriptor: | BARSTAR | | Authors: | Ratnaparkhi, G.S, Varadarajan, R. | | Deposit date: | 1997-12-25 | | Release date: | 1998-04-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Discrepancies between the NMR and X-ray structures of uncomplexed barstar: analysis suggests that packing densities of protein structures determined by NMR are unreliable.
Biochemistry, 37, 1998
|
|
4D0G
 
 | | Structure of Rab14 in complex with Rab-Coupling Protein (RCP) | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RAB11 FAMILY-INTERACTING PROTEIN 1, ... | | Authors: | Lall, P, Khan, A.R. | | Deposit date: | 2014-04-25 | | Release date: | 2015-05-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Function Analyses of the Interactions between Rab11 and Rab14 Small Gtpases with Their Shared Effector Rab Coupling Protein (Rcp).
J.Biol.Chem., 290, 2015
|
|
4FSX
 
 | |
5A9Y
 
 | | Structure of ppGpp BipA | | Descriptor: | GTP-BINDING PROTEIN, GUANOSINE-5',3'-TETRAPHOSPHATE | | Authors: | Kumar, V, Chen, Y, Ero, R, Li, Z, Gao, Y.-G. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5A9X
 
 | | Structure of GDP bound BipA | | Descriptor: | GTP-BINDING PROTEIN, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Kumar, V, Chen, Y, Ero, R, Li, Z, Gao, Y.-G. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5DER
 
 | | RNA oligonucleotide containing (R)-C5'-ME-2'F U | | Descriptor: | RNA oligonucleotide containing (R)-C5'-Me-2'-FU | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2015-08-25 | | Release date: | 2016-06-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Duplex Thermodynamic Stability and Enhanced Nuclease Resistance of 5'-C-Methyl Pyrimidine-Modified Oligonucleotides.
J.Org.Chem., 81, 2016
|
|
4FT4
 
 | |
5D8T
 
 | | RNA octamer containing (S)-5' methyl, 2'-F U. | | Descriptor: | COBALT HEXAMMINE(III), RNA oligonucleotide containing (S)-C5'-Me-2'-FU | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2015-08-17 | | Release date: | 2016-06-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Basis of Duplex Thermodynamic Stability and Enhanced Nuclease Resistance of 5'-C-Methyl Pyrimidine-Modified Oligonucleotides.
J.Org.Chem., 81, 2016
|
|
4FT2
 
 | |
1IJC
 
 | | Solution Structure of Bucandin, a Neurotoxin from the Venom of the Malayan Krait | | Descriptor: | bucandin | | Authors: | Torres, A.M, Kini, R.M, Nirthanan, S, Kuchel, P.W. | | Deposit date: | 2001-04-25 | | Release date: | 2001-12-21 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR structure of bucandin, a neurotoxin from the venom of the Malayan krait (Bungarus candidus).
Biochem.J., 360, 2001
|
|
2KQT
 
 | | Solid-state NMR structure of the M2 transmembrane peptide of the influenza A virus in DMPC lipid bilayers bound to deuterated amantadine | | Descriptor: | (3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-amine, M2 protein | | Authors: | Cady, S.D, Schmidt-Rohr, K, Wang, J, Soto, C.S, DeGrado, W.F, Hong, M. | | Deposit date: | 2009-11-18 | | Release date: | 2010-02-09 | | Last modified: | 2024-05-08 | | Method: | SOLID-STATE NMR | | Cite: | Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers
Nature, 463, 2010
|
|
4OUC
 
 | | Structure of human haspin in complex with histone H3 substrate | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, 1,2-ETHANEDIOL, Histone H3.2, ... | | Authors: | Chaikuad, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-02-15 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Modulation of the chromatin phosphoproteome by the haspin protein kinase.
Mol Cell Proteomics, 13, 2014
|
|
