5XOM
 
 | | Hydra Fam20 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycosaminoglycan xylosylkinase | | Authors: | Xiao, J, Zhang, H. | | Deposit date: | 2017-05-29 | | Release date: | 2018-04-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and evolution of the Fam20 kinases
Nat Commun, 9, 2018
|
|
5YH0
 
 | | The structure of DrFam20C1 | | Descriptor: | DrFam20C1 | | Authors: | Zhang, H, Xiao, J. | | Deposit date: | 2017-09-27 | | Release date: | 2018-04-11 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Structure and evolution of the Fam20 kinases
Nat Commun, 9, 2018
|
|
5YJ5
 
 | |
5YMY
 
 | | The structure of the complex between Rpn13 and K48-diUb | | Descriptor: | Proteasomal ubiquitin receptor ADRM1, Ubiquitin | | Authors: | Liu, Z, Dong, X, Gong, Z, Yi, H.W, Liu, K, Yang, J, Zhang, W.P, Tang, C. | | Deposit date: | 2017-10-22 | | Release date: | 2019-03-13 | | Last modified: | 2019-04-24 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of K48-linked Ub chain by proteasomal receptor Rpn13.
Cell Discov, 5, 2019
|
|
5WJQ
 
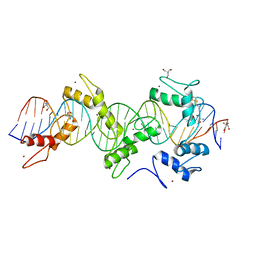 | | mouseZFP568-ZnF2-11 in complex with DNA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA (28-MER), ZINC ION, ... | | Authors: | Patel, A, Cheng, X. | | Deposit date: | 2017-07-24 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | DNA Conformation Induces Adaptable Binding by Tandem Zinc Finger Proteins.
Cell, 173, 2018
|
|
5YH2
 
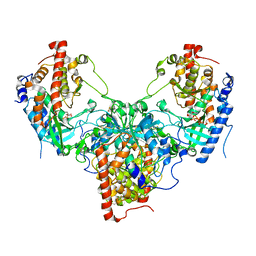 | | The structure of DrFam20C1 and hFam20A complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Family with sequence similarity 20, member Ca, ... | | Authors: | Zhang, H, Xiao, J. | | Deposit date: | 2017-09-27 | | Release date: | 2018-04-11 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Structure and evolution of the Fam20 kinases
Nat Commun, 9, 2018
|
|
5YJ4
 
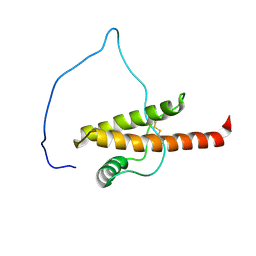 | |
3HQF
 
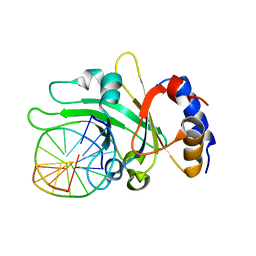 | | Crystal structure of restriction endonuclease EcoRII N-terminal effector-binding domain in complex with cognate DNA | | Descriptor: | 5'-D(*CP*GP*CP*CP*AP*GP*GP*GP*C)-3', 5'-D(*GP*CP*CP*CP*TP*GP*GP*CP*G)-3', Restriction endonuclease | | Authors: | Golovenko, D, Manakova, E, Grazulis, S, Tamulaitiene, G, Siksnys, V. | | Deposit date: | 2009-06-06 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural mechanisms for the 5'-CCWGG sequence recognition by the N- and C-terminal domains of EcoRII.
Nucleic Acids Res., 37, 2009
|
|
3HQG
 
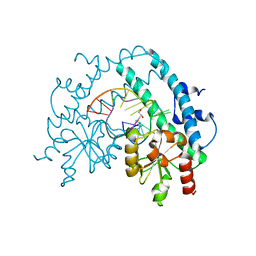 | | Crystal structure of restriction endonuclease EcoRII catalytic C-terminal domain in complex with cognate DNA | | Descriptor: | 5'-D(*TP*AP*GP*CP*CP*TP*GP*GP*TP*CP*GP*A)-3', 5'-D(*TP*CP*GP*AP*CP*CP*AP*GP*GP*CP*TP*A)-3', GLYCEROL, ... | | Authors: | Golovenko, D, Manakova, E, Grazulis, S, Tamulaitiene, G, Siksnys, V. | | Deposit date: | 2009-06-06 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural mechanisms for the 5'-CCWGG sequence recognition by the N- and C-terminal domains of EcoRII.
Nucleic Acids Res., 37, 2009
|
|
5Z7A
 
 | | Crystal structure of NDP52 SKICH region | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Calcium-binding and coiled-coil domain-containing protein 2, GLYCEROL, ... | | Authors: | Pan, L.F, Fu, T, Liu, J.P, Xie, X.Q. | | Deposit date: | 2018-01-27 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Mechanistic insights into the interactions of NAP1 with the SKICH domains of NDP52 and TAX1BP1
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5Z7L
 
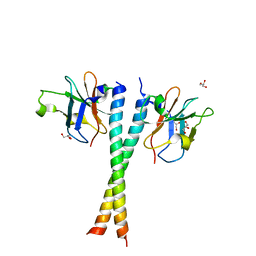 | | Crystal structure of NDP52 SKICH region in complex with NAP1 | | Descriptor: | 5-azacytidine-induced protein 2, Calcium-binding and coiled-coil domain-containing protein 2, GLYCEROL | | Authors: | Fu, T, Pan, L.F. | | Deposit date: | 2018-01-29 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Mechanistic insights into the interactions of NAP1 with the SKICH domains of NDP52 and TAX1BP1
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7VZT
 
 | | A human neutralizing antibody targeting SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GH12-Heavy, GH12-LIGHT, ... | | Authors: | Wang, F.Z, Wang, Y, Tan, X.W, Shi, R, Yan, J.H. | | Deposit date: | 2021-11-16 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | GH12, glycosylation function
To Be Published
|
|
5ZAT
 
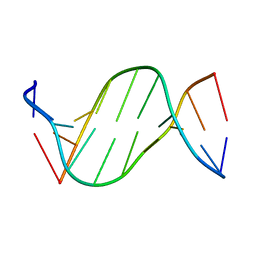 | | Crystal structure of 5-carboxylcytosine containing decamer dsDNA | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*(CAC)P*GP*CP*TP*GP*G)-3') | | Authors: | Fu, T.R, Zhang, L. | | Deposit date: | 2018-02-08 | | Release date: | 2019-02-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Thymine DNA glycosylase recognizes the geometry alteration of minor grooves induced by 5-formylcytosine and 5-carboxylcytosine.
Chem Sci, 10, 2019
|
|
5XOO
 
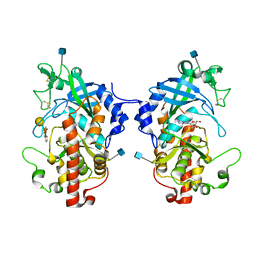 | | The structure of hydra Fam20 with sugar | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE, Glycosaminoglycan xylosylkinase, ... | | Authors: | Zhang, H, Xiao, J. | | Deposit date: | 2017-05-29 | | Release date: | 2018-04-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure and evolution of the Fam20 kinases
Nat Commun, 9, 2018
|
|
5XE5
 
 | | Discovery and structural analysis of a phloretin hydrolase from the opportunistic pathogen Mycobacterium abscessus | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, phloretin hydrolase | | Authors: | He, Y.X, Han, J.T, Jia, W.J, Zhang, Z. | | Deposit date: | 2017-04-01 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Discovery and structural analysis of a phloretin hydrolase from the opportunistic human pathogen Mycobacterium abscessus.
FEBS J., 2019
|
|
5ZAS
 
 | | Crystal structure of 5-formylcytosine containing decamer dsDNA | | Descriptor: | BICARBONATE ION, DNA (5'-D(*CP*CP*AP*GP*(5FC)P*GP*CP*TP*GP*G)-3') | | Authors: | Fu, T.R, Zhang, L. | | Deposit date: | 2018-02-08 | | Release date: | 2019-02-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Thymine DNA glycosylase recognizes the geometry alteration of minor grooves induced by 5-formylcytosine and 5-carboxylcytosine.
Chem Sci, 10, 2019
|
|
7W2P
 
 | | Tudor domain of SMN in complex with a small molecule | | Descriptor: | 2-[(4-fluorophenyl)methyl]-2-azatricyclo[7.3.0.0^{3,7}]dodeca-1(9),3(7)-dien-8-imine, MAGNESIUM ION, Survival motor neuron protein, ... | | Authors: | Li, W, Arrowsmith, C.H, Edwards, A.M, Liu, Y, Min, J. | | Deposit date: | 2021-11-24 | | Release date: | 2022-10-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | A small molecule antagonist of SMN disrupts the interaction between SMN and RNAP II.
Nat Commun, 13, 2022
|
|
7W30
 
 | | Tudor domain of SMN in complex with a small molecule | | Descriptor: | 1,2-dimethylquinolin-4-imine, Survival motor neuron protein, UNKNOWN ATOM OR ION | | Authors: | Li, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Liu, Y, Min, J. | | Deposit date: | 2021-11-24 | | Release date: | 2022-10-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A small molecule antagonist of SMN disrupts the interaction between SMN and RNAP II.
Nat Commun, 13, 2022
|
|
8IZB
 
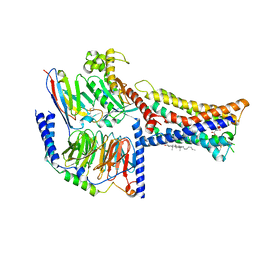 | | Lysophosphatidylserine receptor GPR174-Gs complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Gong, W, Liu, G, Li, X, Zhang, X. | | Deposit date: | 2023-04-06 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis for ligand recognition and signaling of the lysophosphatidylserine receptors GPR34 and GPR174.
Plos Biol., 21, 2023
|
|
8IZF
 
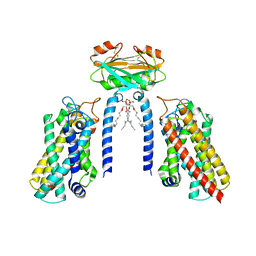 | | Cryo-EM structure of the Lac1-Lip1 (Lip1-S74F) complex | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, Ceramide synthase LAC1, Ceramide synthase subunit LIP1 | | Authors: | Xie, T, Fang, Q, Gong, X. | | Deposit date: | 2023-04-07 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structure and mechanism of a eukaryotic ceramide synthase complex.
Embo J., 42, 2023
|
|
8IZD
 
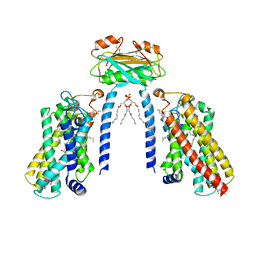 | | Cryo-EM structure of the C26-CoA-bound Lac1-Lip1 complex | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, Ceramide synthase LAC1, Ceramide synthase subunit LIP1, ... | | Authors: | Xie, T, Fang, Q, Gong, X. | | Deposit date: | 2023-04-07 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structure and mechanism of a eukaryotic ceramide synthase complex.
Embo J., 42, 2023
|
|
8IWT
 
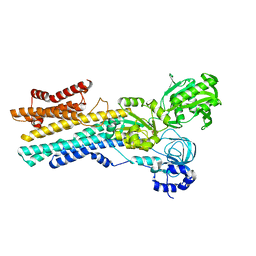 | | hSPCA1 in the early E2P state | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Calcium-transporting ATPase type 2C member 1, MAGNESIUM ION | | Authors: | Liu, Z.M, Wu, M.Q, Wu, C. | | Deposit date: | 2023-03-31 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structure and transport mechanism of the human calcium pump SPCA1.
Cell Res., 33, 2023
|
|
8IWR
 
 | | hSPCA1 in the CaE1-ATP state | | Descriptor: | CALCIUM ION, Calcium-transporting ATPase type 2C member 1, MAGNESIUM ION, ... | | Authors: | Liu, Z.M, Wu, M.Q, Wu, C. | | Deposit date: | 2023-03-31 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structure and transport mechanism of the human calcium pump SPCA1.
Cell Res., 33, 2023
|
|
8IWP
 
 | | hSPCA1 in the CaE1 state | | Descriptor: | CALCIUM ION, Calcium-transporting ATPase type 2C member 1 | | Authors: | Liu, Z.M, Wu, M.Q, Wu, C. | | Deposit date: | 2023-03-30 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structure and transport mechanism of the human calcium pump SPCA1.
Cell Res., 33, 2023
|
|
8IWW
 
 | | hSPCA1 in the CaE1P-ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, Calcium-transporting ATPase type 2C member 1, ... | | Authors: | Liu, Z.M, Wu, M.Q, Wu, C. | | Deposit date: | 2023-03-31 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structure and transport mechanism of the human calcium pump SPCA1.
Cell Res., 33, 2023
|
|
