3E51
 
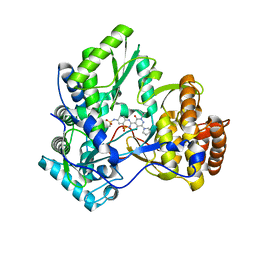 | | Crystal structure of HCV NS5B polymerase with a novel pyridazinone inhibitor | | Descriptor: | N-{3-[5-hydroxy-2-(3-methylbutyl)-3-oxo-6-pyrrolidin-1-yl-2,3-dihydropyridazin-4-yl]-1,1-dioxido-2H-1,2,4-benzothiadiazin-7-yl}methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Han, Q, Showalter, R.E, Zhao, Q, Kissinger, C.R. | | Deposit date: | 2008-08-12 | | Release date: | 2009-08-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Novel HCV NS5B polymerase inhibitors derived from 4-(1',1'-dioxo-1',4'-dihydro-1'lambda(6)-benzo[1',2',4']thiadiazin-3'-yl)-5-hydroxy-2H-pyridazin-3-ones. Part 5: Exploration of pyridazinones containing 6-amino-substituents.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
1WOF
 
 | | Crystal Structure Of SARS-CoV Mpro in Complex with an Inhibitor N1 | | Descriptor: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]-L-ALANYL-L-VALYL-N~1~-((1S)-4-ETHOXY-4-OXO-1-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Yang, H, Bartlam, M, Xue, X, Yang, K, Liang, W, Rao, Z. | | Deposit date: | 2004-08-18 | | Release date: | 2005-08-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design of Wide-Spectrum Inhibitors Targeting Coronavirus Main Proteases.
Plos Biol., 3, 2005
|
|
1VJI
 
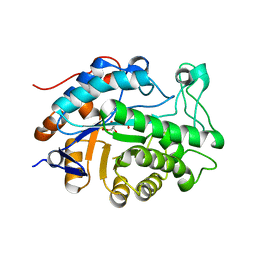 | | Gene Product of At1g76680 from Arabidopsis thaliana | | Descriptor: | 12-oxophytodienoate reductase (OPR1), FLAVIN MONONUCLEOTIDE | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Johnson, K.A, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-02-24 | | Release date: | 2004-03-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | X-ray structure of Arabidopsis At1g77680, 12-oxophytodienoate reductase isoform 1.
Proteins, 61, 2005
|
|
7DR6
 
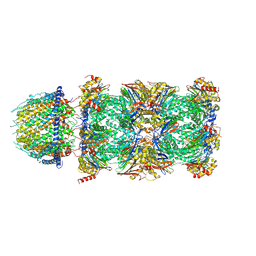 | | PA28alpha-beta in complex with immunoproteasome | | Descriptor: | Proteasome activator complex subunit 1, Proteasome activator complex subunit 2, Proteasome subunit alpha type-1, ... | | Authors: | Cong, Y, Xu, C. | | Deposit date: | 2020-12-26 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM of mammalian PA28 alpha beta-iCP immunoproteasome reveals a distinct mechanism of proteasome activation by PA28 alpha beta.
Nat Commun, 12, 2021
|
|
7DR7
 
 | | bovine 20S immunoproteasome | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2020-12-26 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM of mammalian PA28 alpha beta-iCP immunoproteasome reveals a distinct mechanism of proteasome activation by PA28 alpha beta.
Nat Commun, 12, 2021
|
|
7DRW
 
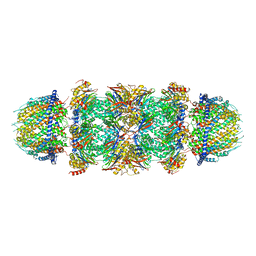 | | Bovine 20S immunoproteasome in complex with two human PA28alpha-beta activators | | Descriptor: | Proteasome activator complex subunit 1, Proteasome activator complex subunit 2, Proteasome subunit alpha type-1, ... | | Authors: | Cong, Y, Xu, C. | | Deposit date: | 2020-12-29 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM of mammalian PA28 alpha beta-iCP immunoproteasome reveals a distinct mechanism of proteasome activation by PA28 alpha beta.
Nat Commun, 12, 2021
|
|
2Q8T
 
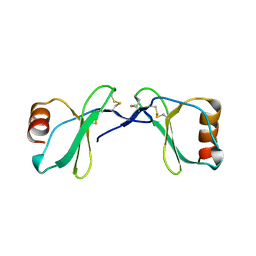 | | Crystal Structure of the CC chemokine CCL14 | | Descriptor: | CCL14 | | Authors: | Blain, K.Y. | | Deposit date: | 2007-06-11 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural and Functional Characterization of CC Chemokine CCL14
Biochemistry, 46, 2007
|
|
2Q6D
 
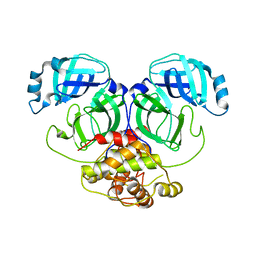 | | Crystal structure of infectious bronchitis virus (IBV) main protease | | Descriptor: | Infectious bronchitis virus (IBV) main protease | | Authors: | Xue, X.Y, Yang, H.T, Xue, F, Bartlam, M, Rao, Z.H. | | Deposit date: | 2007-06-04 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design.
J.Virol., 82, 2008
|
|
2Q6G
 
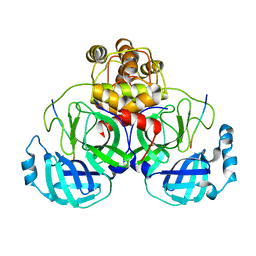 | | Crystal structure of SARS-CoV main protease H41A mutant in complex with an N-terminal substrate | | Descriptor: | Polypeptide chain, severe acute respiratory syndrome coronavirus (SARS-CoV) | | Authors: | Xue, X.Y, Yang, H.T, Xue, F, Bartlam, M, Rao, Z.H. | | Deposit date: | 2007-06-05 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design.
J.Virol., 82, 2008
|
|
2Q6F
 
 | | Crystal structure of infectious bronchitis virus (IBV) main protease in complex with a Michael acceptor inhibitor N3 | | Descriptor: | Infectious bronchitis virus (IBV) main protease, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Xue, X.Y, Yang, H.T, Xue, F, Bartlam, M, Rao, Z.H. | | Deposit date: | 2007-06-05 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design.
J.Virol., 82, 2008
|
|
2Q8R
 
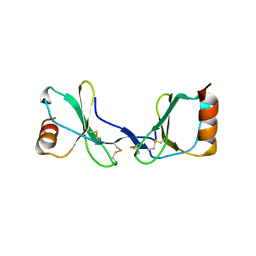 | |
6WI7
 
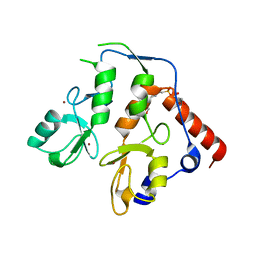 | | RING1B-BMI1 fusion in closed conformation | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, E3 ubiquitin-protein ligase RING2, Polycomb complex protein BMI-1 chimera, ... | | Authors: | Cho, H.J, Cierpicki, T. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Small-molecule inhibitors targeting Polycomb repressive complex 1 RING domain.
Nat.Chem.Biol., 17, 2021
|
|
6WI8
 
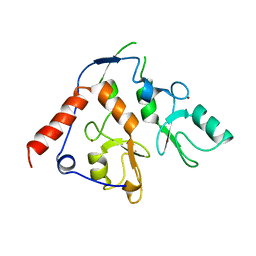 | |
6XFV
 
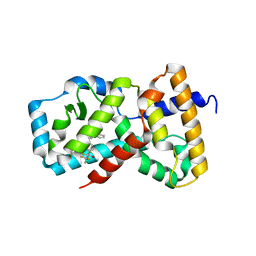 | | CRYSTAL STRUCTURE OF RAR-RELATED ORPHAN RECEPTOR C IN COMPLEX WITH A NOVEL INVERSE AGONIST | | Descriptor: | 1-(4-{(3S,4S)-4-[4-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)phenyl]-3-methyl-3-phenylpyrrolidine-1-carbonyl}piperidin-1-yl)ethan-1-one, Nuclear receptor ROR-gamma | | Authors: | Sack, J.S. | | Deposit date: | 2020-06-16 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of (3S,4S)-3-methyl-3-(4-fluorophenyl)-4-(4-(1,1,1,3,3,3-hexafluoro-2-hydroxyprop-2-yl)phenyl)pyrrolidines as novel ROR gamma t inverse agonists.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6LF5
 
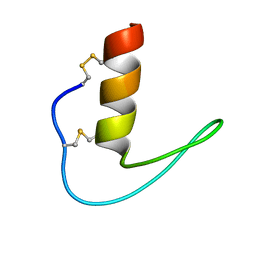 | | The solution structure of ShSPI | | Descriptor: | ShSPI | | Authors: | Luan, N, Rong, M.Q, Liu, J.X, Lai, R. | | Deposit date: | 2019-11-29 | | Release date: | 2020-12-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Identification and Characterization of ShSPI, a Kazal-Type Elastase Inhibitor from the Venom of Scolopendra Hainanum .
Toxins, 11, 2019
|
|
6KOW
 
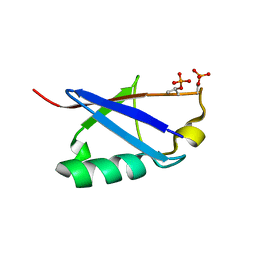 | |
6J2W
 
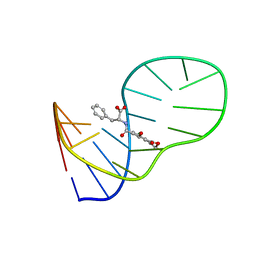 | | The structure of OBA3-OTA complex | | Descriptor: | DNA (5'-D(*CP*GP*GP*GP*GP*CP*GP*AP*AP*GP*CP*GP*GP*GP*TP*CP*CP*CP*G)-3'), N-[(3R)-5-chloro-8-hydroxy-3-methyl-1-oxo-3,4-dihydro-1H-2-benzopyran-7-carbonyl]-D-phenylalanine | | Authors: | Xu, G.H, Li, C.G. | | Deposit date: | 2019-01-03 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure-guided post-SELEX optimization of an ochratoxin A aptamer.
Nucleic Acids Res., 47, 2019
|
|
4GAX
 
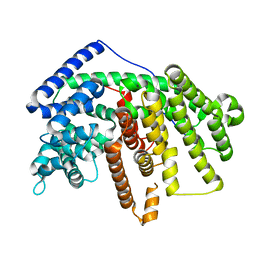 | | Crystal Structure of an alpha-Bisabolol synthase mutant | | Descriptor: | Amorpha-4,11-diene synthase | | Authors: | Li, J, Peng, Z. | | Deposit date: | 2012-07-26 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9948 Å) | | Cite: | Rational engineering of plasticity residues of sesquiterpene synthases from Artemisia annua: product specificity and catalytic efficiency.
Biochem.J., 451, 2013
|
|
5CDI
 
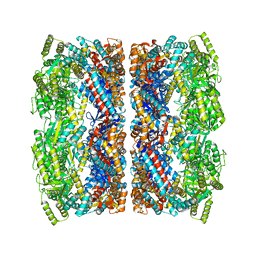 | | Chloroplast chaperonin 60b1 of Chlamydomonas | | Descriptor: | Chaperonin 60B1 | | Authors: | Zhang, S, Zhou, H, Yu, F, Gao, F, He, J, Liu, C. | | Deposit date: | 2015-07-04 | | Release date: | 2016-05-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.807 Å) | | Cite: | Structural insight into the cooperation of chloroplast chaperonin subunits
Bmc Biol., 14, 2016
|
|
5CMA
 
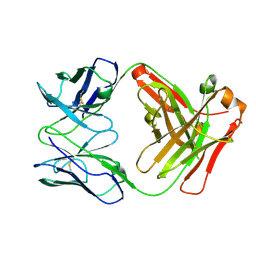 | | Anti-B7-H3 monoclonal antibody ch8H9 Fab fragment | | Descriptor: | Antibody ch8H9 Fab heavy chain, Antibody ch8H9 Fab light chain | | Authors: | Ahmed, M, Goldgur, Y, Cheng, M, Cheung, N.K. | | Deposit date: | 2015-07-16 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Humanized Affinity-matured Monoclonal Antibody 8H9 Has Potent Antitumor Activity and Binds to FG Loop of Tumor Antigen B7-H3.
J.Biol.Chem., 290, 2015
|
|
4FJQ
 
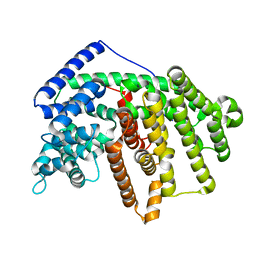 | | Crystal Structure of an alpha-Bisabolol synthase | | Descriptor: | Amorpha-4,11-diene synthase | | Authors: | Jianxu, L, Peng, Z. | | Deposit date: | 2012-06-12 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.0001 Å) | | Cite: | Rational engineering of plasticity residues of sesquiterpene synthases from Artemisia annua: product specificity and catalytic efficiency.
Biochem.J., 451, 2013
|
|
1H66
 
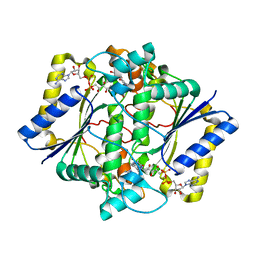 | | CRYSTAL STRUCTURE OF HUMAN NAD[P]H-QUINONE OXIDOREDUCTASE CO WITH 2,5-diaziridinyl-3-hydroxyl-6-methyl-1,4-benzoquinone | | Descriptor: | 2,5-DIAZIRIDIN-1-YL-3-(HYDROXYMETHYL)-6-METHYLCYCLOHEXA-2,5-DIENE-1,4-DIONE, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H DEHYDROGENASE [QUINONE] 1 | | Authors: | Faig, M, Bianchet, M.A, Winski, S, Ross, D, Amzel, L.M. | | Deposit date: | 2001-06-06 | | Release date: | 2001-09-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Development of Anticancer Drugs: Complexes of Nad(P)H:Quinone Oxidoreductase 1 with Chemotherapeutic Quinones
Structure, 9, 2001
|
|
1H69
 
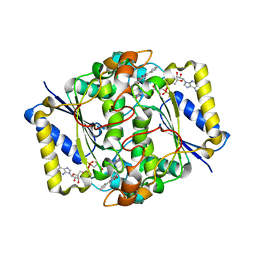 | | CRYSTAL STRUCTURE OF HUMAN NAD[P]H-QUINONE OXIDOREDUCTASE CO WITH 2,3,5,6,TETRAMETHYL-P-BENZOQUINONE (DUROQUINONE) AT 2.5 ANGSTROM RESOLUTION | | Descriptor: | 3-(HYDROXYMETHYL)-1-METHYL-5-(2-METHYLAZIRIDIN-1-YL)-2-PHENYL-1H-INDOLE-4,7-DIONE, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H DEHYDROGENASE [QUINONE] 1 | | Authors: | Faig, M, Bianchet, M.A, Chen, S, Winski, S, Ross, D, Amzel, L.M. | | Deposit date: | 2001-06-08 | | Release date: | 2001-09-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure-Based Development of Anticancer Drugs: Complexes of Nad(P)H:Quinone Oxidoreductase 1 with Chemotherapeutic Quinones
Structure, 9, 2001
|
|
1GG5
 
 | | CRYSTAL STRUCTURE OF A COMPLEX OF HUMAN NAD[P]H-QUINONE OXIDOREDUCTASE AND A CHEMOTHERAPEUTIC DRUG (E09) AT 2.5 A RESOLUTION | | Descriptor: | 3-HYDROXYMETHYL-5-AZIRIDINYL-1METHYL-2-[1H-INDOLE-4,7-DIONE]-PROPANOL, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H DEHYDROGENASE [QUINONE] 1 | | Authors: | Faig, M, Bianchet, M.A, Winski, S, Hargreaves, R, Moody, C.J, Hudnott, A.R, Ross, D, Amzel, L.M. | | Deposit date: | 2000-07-12 | | Release date: | 2001-09-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based development of anticancer drugs: complexes of NAD(P)H:quinone oxidoreductase 1 with chemotherapeutic quinones.
Structure, 9, 2001
|
|
6INC
 
 | | Crystal structure of an acetolactate decarboxylase from Klebsiella pneumoniae | | Descriptor: | 1,2-ETHANEDIOL, Alpha-acetolactate decarboxylase, CHLORIDE ION, ... | | Authors: | Wu, W, Zhang, Q, Bartlam, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Structural characterization of an acetolactate decarboxylase from Klebsiella pneumoniae
Biochem. Biophys. Res. Commun., 509, 2019
|
|
