5YM8
 
 | |
5YM6
 
 | |
5Y4L
 
 | | PRRSV nsp4 | | Descriptor: | Non-structural protein | | Authors: | Shi, Y.J, Gang, Y, Peng, G.Q. | | Deposit date: | 2017-08-04 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Identification of two antiviral inhibitors targeting 3C-like serine/3C-like protease of porcine reproductive and respiratory syndrome virus and porcine epidemic diarrhea virus.
Vet. Microbiol., 213, 2018
|
|
3QK5
 
 | | Crystal structure of fatty acid amide hydrolase with small molecule inhibitor | | Descriptor: | (3-{(3R)-1-[4-(1-benzothiophen-2-yl)pyrimidin-2-yl]piperidin-3-yl}-2-methyl-1H-pyrrolo[2,3-b]pyridin-1-yl)acetonitrile, 1,2-ETHANEDIOL, Fatty-acid amide hydrolase 1, ... | | Authors: | Min, X, Walker, N.P.C, Wang, Z. | | Deposit date: | 2011-01-31 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of potent, noncovalent fatty acid amide hydrolase (FAAH) inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4QQN
 
 | | Protein arginine methyltransferase 3 in complex with compound MTV044246 | | Descriptor: | 1-{2-[1-(aminomethyl)cyclohexyl]ethyl}-3-isoquinolin-6-ylurea, CHLORIDE ION, GLYCEROL, ... | | Authors: | Dong, A, Dobrovetsky, E, Tempel, W, He, H, Zhao, K, Smil, D, Landon, M, Luo, X, Chen, Z, Dai, M, Yu, Z, Lin, Y, Zhang, H, Zhao, K, Schapira, M, Brown, P.J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-27 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Discovery of Potent and Selective Allosteric Inhibitors of Protein Arginine Methyltransferase 3 (PRMT3).
J. Med. Chem., 61, 2018
|
|
8SYC
 
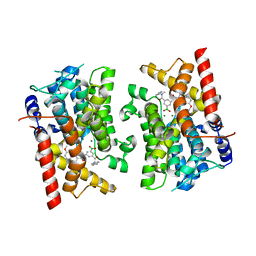 | | Crystal structure of PDE3B in complex with GSK4394835A | | Descriptor: | MAGNESIUM ION, [3-[(4,7-dimethoxyquinolin-2-yl)carbonylamino]-5-[methyl-(phenylmethyl)carbamoyl]phenyl]-oxidanyl-oxidanylidene-boron, cGMP-inhibited 3',5'-cyclic phosphodiesterase 3B | | Authors: | Concha, N.O, Nolte, R. | | Deposit date: | 2023-05-25 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery and SAR Study of Boronic Acid-Based Selective PDE3B Inhibitors from a Novel DNA-Encoded Library.
J.Med.Chem., 67, 2024
|
|
6LKX
 
 | | The structure of PRRSV helicase | | Descriptor: | CITRIC ACID, GLYCEROL, RNA-dependent RNA polymerase, ... | | Authors: | Shi, Y.J, Tong, X.H, Peng, G.Q. | | Deposit date: | 2019-12-20 | | Release date: | 2020-05-27 | | Last modified: | 2021-06-09 | | Method: | X-RAY DIFFRACTION (2.998 Å) | | Cite: | Structural Characterization of the Helicase nsp10 Encoded by Porcine Reproductive and Respiratory Syndrome Virus.
J.Virol., 94, 2020
|
|
6LN0
 
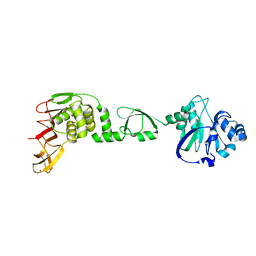 | |
6LU5
 
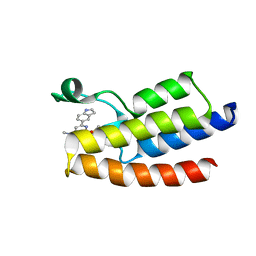 | | Crystal structure of BPTF-BRD with ligand DCBPin5 bound | | Descriptor: | 6-(1H-indol-5-yl)-N-methyl-2-methylsulfonyl-pyrimidin-4-amine, Nucleosome-remodeling factor subunit BPTF | | Authors: | Lu, T, Lu, H.B, Wang, J, Lin, H, Lu, W, Luo, C. | | Deposit date: | 2020-01-25 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.86527729 Å) | | Cite: | Discovery and Optimization of Small-Molecule Inhibitors for the BPTF Bromodomains Proteins
To Be Published
|
|
6LU6
 
 | | Crystal structure of BPTF-BRD with ligand DCBPin5-2 bound | | Descriptor: | 6-[1-[3-(dimethylamino)propyl]indol-5-yl]-2-methylsulfonyl-N-propyl-pyrimidin-4-amine, Nucleosome-remodeling factor subunit BPTF | | Authors: | Lu, T, Lu, H.B, Wang, J, Lin, H, Lu, W, Luo, C. | | Deposit date: | 2020-01-26 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.970063 Å) | | Cite: | Discovery and Optimization of Small-Molecule Inhibitors for the BPTF Bromodomains Proteins
To Be Published
|
|
4XFQ
 
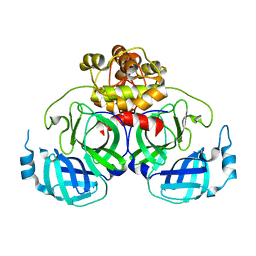 | | Crystal Structure Basis for PEDV 3C Like Protease | | Descriptor: | PEDV main protease | | Authors: | Ye, G, Fu, Z.F, Peng, G.Q. | | Deposit date: | 2014-12-28 | | Release date: | 2016-01-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for the dimerization and substrate recognition specificity of porcine epidemic diarrhea virus 3C-like protease.
Virology, 494, 2016
|
|
5HIY
 
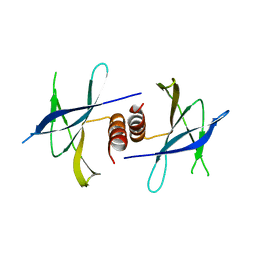 | | Crystal structure of PEDV NSP9 Mutant-C59A | | Descriptor: | Non-structural protein 9 | | Authors: | Deng, F, Peng, G. | | Deposit date: | 2016-01-12 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Dimerization of Coronavirus nsp9 with Diverse Modes Enhances Its Nucleic Acid Binding Affinity.
J.Virol., 92, 2018
|
|
4ZUH
 
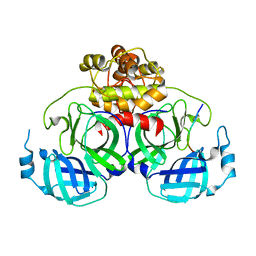 | |
5HIZ
 
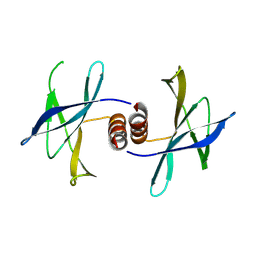 | | The structure of PEDV NSP9 | | Descriptor: | Non-structural protein 9 | | Authors: | Deng, F, Peng, G. | | Deposit date: | 2016-01-12 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Dimerization of Coronavirus nsp9 with Diverse Modes Enhances Its Nucleic Acid Binding Affinity.
J.Virol., 92, 2018
|
|
8K8H
 
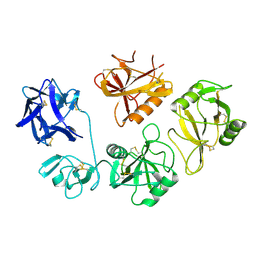 | |
2B5E
 
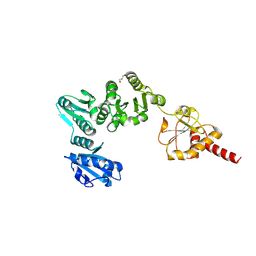 | | Crystal Structure of Yeast Protein Disulfide Isomerase | | Descriptor: | BARIUM ION, GLYCEROL, Protein disulfide-isomerase | | Authors: | Schindelin, H, Tian, G. | | Deposit date: | 2005-09-28 | | Release date: | 2006-01-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of yeast protein disulfide isomerase suggests cooperativity between its active sites.
Cell(Cambridge,Mass.), 124, 2006
|
|
8I4L
 
 | | Capsid structure of the Cyanophage P-SCSP1u | | Descriptor: | The capsid protein(gp 19) of P-SCSP1u | | Authors: | Liu, H, Dang, S. | | Deposit date: | 2023-01-19 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Cryo-EM structure of cyanophage P-SCSP1u offers insights into DNA gating and evolution of T7-like viruses.
Nat Commun, 14, 2023
|
|
8I4M
 
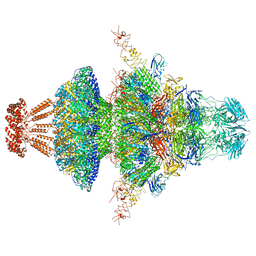 | | Portal-tail complex structure of the Cyanophage P-SCSP1u | | Descriptor: | Adaptor protein(gp22) of the cyanophage P-SCSP1u, Fiber protein(gp 28) of the cyanophage P-SCSP1u, Nozzle protein(gp 23) of the cyanophage P-SCSP1u, ... | | Authors: | Liu, H, Dang, S. | | Deposit date: | 2023-01-19 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Cryo-EM structure of cyanophage P-SCSP1u offers insights into DNA gating and evolution of T7-like viruses.
Nat Commun, 14, 2023
|
|
6IVD
 
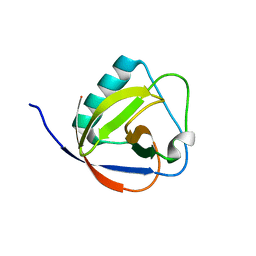 | | TGEV nsp1 mutant - 91-95sg | | Descriptor: | nsp1 mutant protein | | Authors: | Shen, Z, Peng, G.Q. | | Deposit date: | 2018-12-03 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.975 Å) | | Cite: | A conserved region of nonstructural protein 1 from alphacoronaviruses inhibits host gene expression and is critical for viral virulence.
J.Biol.Chem., 294, 2019
|
|
6IVC
 
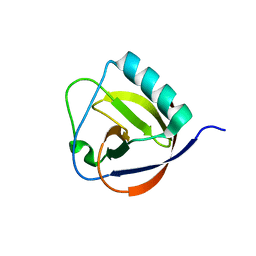 | | The full length of TGEV nsp1 | | Descriptor: | nsp1 protein | | Authors: | Shen, Z, Peng, G.Q. | | Deposit date: | 2018-12-03 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A conserved region of nonstructural protein 1 from alphacoronaviruses inhibits host gene expression and is critical for viral virulence.
J.Biol.Chem., 294, 2019
|
|
3HB9
 
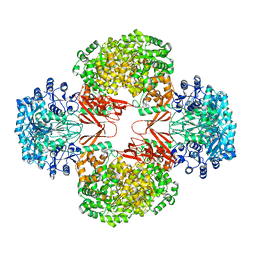 | | Crystal Structure of S. aureus Pyruvate Carboxylase A610T Mutant | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ADENOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Tong, L, Yu, L.P.C. | | Deposit date: | 2009-05-04 | | Release date: | 2009-06-30 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A Symmetrical Tetramer for S. aureus Pyruvate Carboxylase in Complex with Coenzyme A.
Structure, 17, 2009
|
|
3HO8
 
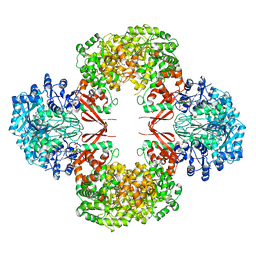 | |
3HBL
 
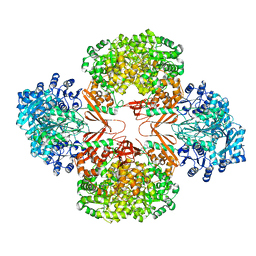 | | Crystal Structure of S. aureus Pyruvate Carboxylase T908A Mutant | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ADENOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Tong, L, Yu, L.P.C. | | Deposit date: | 2009-05-04 | | Release date: | 2009-06-30 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | A Symmetrical Tetramer for S. aureus Pyruvate Carboxylase in Complex with Coenzyme A.
Structure, 17, 2009
|
|
7DHJ
 
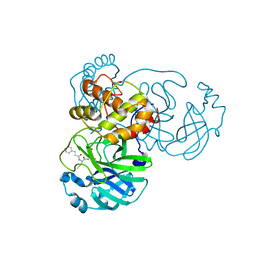 | | The co-crystal structure of SARS-CoV-2 main protease with the peptidomimetic inhibitor (S)-2-cinnamamido-N-((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)pent-4-ynamide | | Descriptor: | (2~{S})-~{N}-[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-2-[[(~{E})-3-phenylprop-2-enoyl]amino]pent-4-ynamide, 3C-like proteinase | | Authors: | Shang, L.Q, Wang, H, Deng, W.L, Xing, S, Wang, Y.X. | | Deposit date: | 2020-11-15 | | Release date: | 2021-11-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | The structure-based design of peptidomimetic inhibitors against SARS-CoV-2 3C like protease as Potent anti-viral drug candidate.
Eur.J.Med.Chem., 238, 2022
|
|
6RM9
 
 | | Crystal structure of the DEAH-box ATPase Prp2 in complex with Spp2 and ADP | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hamann, F, Neumann, P, Ficner, R. | | Deposit date: | 2019-05-06 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural analysis of the intrinsically disordered splicing factor Spp2 and its binding to the DEAH-box ATPase Prp2.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
