7Y6W
 
 | | RRGSGG-AtPRT6 UBR box (I222) | | Descriptor: | E3 ubiquitin-protein ligase PRT6, ZINC ION | | Authors: | Kim, L, Song, H.K. | | Deposit date: | 2022-06-21 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analyses of plant PRT6-UBR box for Cys-Arg/N-degron pathway and insights into the plant submergence resistance
To Be Published
|
|
7Y6Z
 
 | | RLGSGG-AtPRT6 UBR box (I222) | | Descriptor: | E3 ubiquitin-protein ligase PRT6, ZINC ION | | Authors: | Kim, L, Song, H.K. | | Deposit date: | 2022-06-21 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Structural analyses of plant PRT6-UBR box for Cys-Arg/N-degron pathway and insights into the plant submergence resistance
To Be Published
|
|
4H8A
 
 | | Crystal structure of ureidoglycolate dehydrogenase in binary complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Ureidoglycolate dehydrogenase | | Authors: | Rhee, S, Shin, I, Kim, M. | | Deposit date: | 2012-09-22 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural and functional insights into (s)-ureidoglycolate dehydrogenase, a metabolic branch point enzyme in nitrogen utilization.
Plos One, 7, 2012
|
|
7XL9
 
 | | The structure of HucR with urate | | Descriptor: | CHLORIDE ION, Transcriptional regulator, MarR family, ... | | Authors: | Park, S.Y. | | Deposit date: | 2022-04-21 | | Release date: | 2022-07-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | The structure of Deinococcus radiodurans transcriptional regulator HucR retold with the urate bound.
Biochem.Biophys.Res.Commun., 615, 2022
|
|
5WDF
 
 | |
5XJV
 
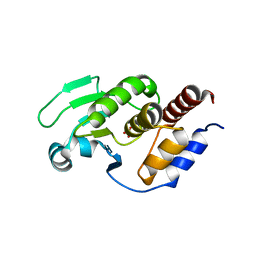 | | Two intermediate states of conformation switch in dual specificity phosphatase 13a | | Descriptor: | Dual specificity protein phosphatase 13 isoform A, PHOSPHATE ION | | Authors: | Wei, C.H, Min, H.G, Chun, H.J, Ryu, S.E. | | Deposit date: | 2017-05-04 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Two intermediate states of the conformational switch in dual specificity phosphatase 13a
Pharmacol. Res., 128, 2018
|
|
6BSI
 
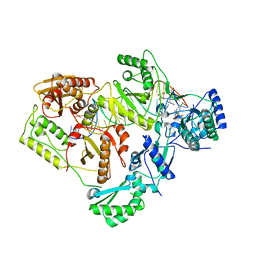 | | Structure of HIV-1 RT complexed with an RNA/DNA hybrid containing the polypurine-tract sequence | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, CALCIUM ION, DNA (5'-D(*GP*TP*TP*TP*TP*TP*CP*TP*TP*TP*TP*GP*TP*TP*AP*TP*TP*GP*TP*GP*GP*CP*C)-3'), ... | | Authors: | Tian, L, Kim, M, Yang, W. | | Deposit date: | 2017-12-03 | | Release date: | 2018-01-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure of HIV-1 reverse transcriptase cleaving RNA in an RNA/DNA hybrid.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BSJ
 
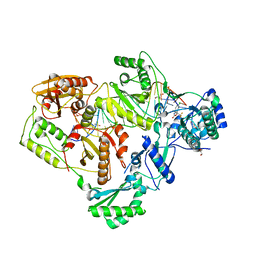 | | Structure of HIV-1 RT complexed with an RNA/DNA hybrid sequence non-preferred for RNA hydrolysis | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, CALCIUM ION, DNA (5'-D(*GP*TP*AP*TP*GP*CP*CP*TP*AP*TP*AP*GP*TP*TP*AP*TP*TP*GP*TP*GP*GP*CP*C)-3'), ... | | Authors: | Tian, L, Kim, M, Yang, W. | | Deposit date: | 2017-12-03 | | Release date: | 2018-01-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure of HIV-1 reverse transcriptase cleaving RNA in an RNA/DNA hybrid.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BSH
 
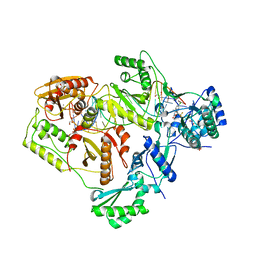 | | Structure of HIV-1 RT complexed with RNA/DNA hybrid in the RNA hydrolysis mode | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, CALCIUM ION, DNA (5'-D(*GP*TP*AP*TP*GP*CP*CP*AP*CP*TP*AP*GP*TP*TP*AP*TP*TP*GP*TP*GP*GP*CP*C)-3'), ... | | Authors: | Tian, L, Kim, M, Yang, W. | | Deposit date: | 2017-12-03 | | Release date: | 2018-01-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.649 Å) | | Cite: | Structure of HIV-1 reverse transcriptase cleaving RNA in an RNA/DNA hybrid.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BSG
 
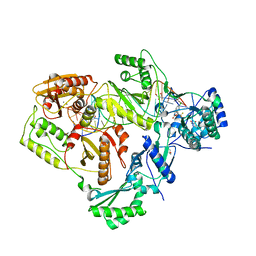 | | Structure of HIV-1 RT complexed with RNA/DNA hybrid in an RNA hydrolysis-off mode | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tian, L, Kim, M, Yang, W. | | Deposit date: | 2017-12-03 | | Release date: | 2018-01-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structure of HIV-1 reverse transcriptase cleaving RNA in an RNA/DNA hybrid.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8DZV
 
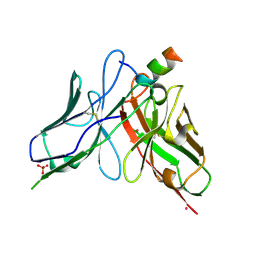 | |
9DUK
 
 | |
9DUL
 
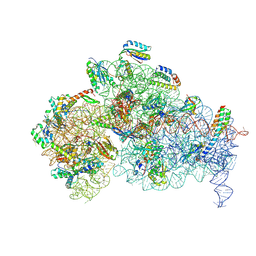 | |
5T8I
 
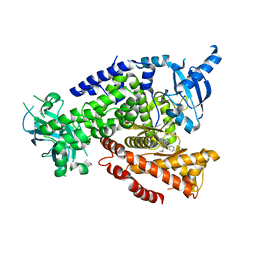 | | PI3Kdelta in complex with the inhibitor GS-9901 | | Descriptor: | 2,4-diamino-6-{[(S)-[5-chloro-8-fluoro-4-oxo-3-(pyridin-3-yl)-3,4-dihydroquinazolin-2-yl](cyclopropyl)methyl]amino}pyrimidine-5-carbonitrile, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Somoza, J.R, Villasenor, A. | | Deposit date: | 2016-09-07 | | Release date: | 2016-12-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Discovery of GS-9901: A Potent, Selective and Metabolically Stable Inhibitor of PI3Kd
To Be Published
|
|
5T7F
 
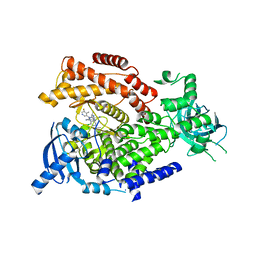 | | PI3Kdelta in complex with the inhibitor GS-643624 | | Descriptor: | 2,4-bis(azanyl)-6-[[(1~{S})-1-[5-chloranyl-3-(5-fluoranyl-4-methyl-pyridin-3-yl)-4-oxidanylidene-quinazolin-2-yl]ethyl]amino]pyrimidine-5-carbonitrile, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Somoza, J.R, Villasenor, A. | | Deposit date: | 2016-09-04 | | Release date: | 2016-12-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Discovery of GS-9901: A Potent, Selective and Metabolically Stable Inhibitor of PI3Kd
To Be Published
|
|
8FRR
 
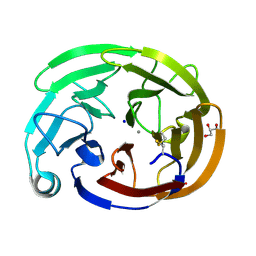 | | Wild-type myocilin olfactomedin domain | | Descriptor: | CALCIUM ION, GLYCEROL, Myocilin, ... | | Authors: | Ma, M.T, Lieberman, R.L, Huard, D.J.E. | | Deposit date: | 2023-01-08 | | Release date: | 2024-01-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Competition between inside-out unfolding and pathogenic aggregation in an amyloid-forming beta-propeller.
Nat Commun, 15, 2024
|
|
3ADE
 
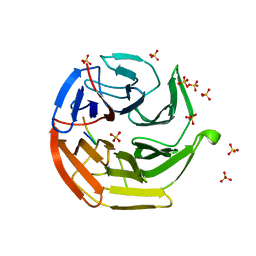 | | Crystal Structure of Keap1 in Complex with Sequestosome-1/p62 | | Descriptor: | Kelch-like ECH-associated protein 1, SULFATE ION, Sequestosome-1 | | Authors: | Kurokawa, H, Yamamoto, M. | | Deposit date: | 2010-01-19 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1
Nat.Cell Biol., 12, 2010
|
|
7XL8
 
 | | Human Cx36/GJD2 (N-terminal deletion mutant) gap junction channel in soybean lipids (D6 symmetry) | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction delta-2 protein | | Authors: | Lee, S.N, Cho, H.J, Jeong, H, Ryu, B, Lee, H.J, Lee, H.H, Woo, J.S. | | Deposit date: | 2022-04-21 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of human Cx36/GJD2 neuronal gap junction channel.
Nat Commun, 14, 2023
|
|
7XNV
 
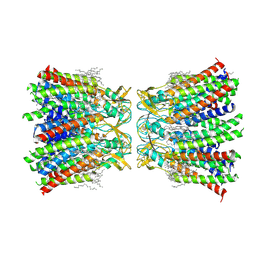 | | Structurally hetero-junctional human Cx36/GJD2 gap junction channel in soybean lipids (C6 symmetry) | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction delta-2 protein | | Authors: | Lee, S.N, Cho, H.J, Jeong, H, Ryu, B, Lee, H.J, Lee, H.H, Woo, J.S. | | Deposit date: | 2022-04-29 | | Release date: | 2023-03-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of human Cx36/GJD2 neuronal gap junction channel.
Nat Commun, 14, 2023
|
|
7XKT
 
 | | Human Cx36/GJD2 (BRIL-fused mutant) gap junction channel in detergents at 2.2 Angstroms resolution | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Cho, H.J, Lee, S.N, Jeong, H, Ryu, B, Lee, H.J, Woo, J.S, Lee, H.H. | | Deposit date: | 2022-04-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Cryo-EM structures of human Cx36/GJD2 neuronal gap junction channel.
Nat Commun, 14, 2023
|
|
7XKK
 
 | | Human Cx36/GJD2 gap junction channel in detergents | | Descriptor: | Gap junction delta-2 protein | | Authors: | Lee, S.N, Cho, H.J, Jeong, H, Ryu, B, Lee, H.J, Lee, H.H, Woo, J.S. | | Deposit date: | 2022-04-19 | | Release date: | 2023-04-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of human Cx36/GJD2 neuronal gap junction channel.
Nat Commun, 14, 2023
|
|
8HKP
 
 | | Structurally hetero-junctional human Cx36/GJD2 gap junction channel in detergents (C6 symmetry) | | Descriptor: | Gap junction delta-2 protein, Lauryl Maltose Neopentyl Glycol | | Authors: | Lee, S.N, Cho, H.J, Jeong, H, Ryu, B, Lee, H.J, Lee, H.H, Woo, J.S. | | Deposit date: | 2022-11-27 | | Release date: | 2023-03-22 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of human Cx36/GJD2 neuronal gap junction channel.
Nat Commun, 14, 2023
|
|
3VRN
 
 | | Crystal structure of the tyrosine kinase binding domain of Cbl-c | | Descriptor: | CALCIUM ION, Signal transduction protein CBL-C | | Authors: | Takeshita, K, Tezuka, T, Isozaki, Y, Yamashita, E, Suzuki, M, Yamanashi, Y, Yamamoto, T, Nakagawa, A. | | Deposit date: | 2012-04-13 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural flexibility regulates phosphopeptide-binding activity of the tyrosine kinase binding domain of Cbl-c
J.Biochem., 152, 2012
|
|
3VRR
 
 | | Crystal structure of the tyrosine kinase binding domain of Cbl-c (PL mutant) in complex with phospho-EGFR peptide | | Descriptor: | CALCIUM ION, Epidermal growth factor receptor, Signal transduction protein CBL-C | | Authors: | Takeshita, K, Tezuka, T, Isozaki, Y, Yamashita, E, Suzuki, M, Yamanashi, Y, Yamamoto, T, Nakagawa, A. | | Deposit date: | 2012-04-13 | | Release date: | 2013-03-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural flexibility regulates phosphopeptide-binding activity of the tyrosine kinase binding domain of Cbl-c.
J.Biochem., 152, 2012
|
|
3VRQ
 
 | | Crystal structure of the tyrosine kinase binding domain of Cbl-c (PL mutant) | | Descriptor: | CALCIUM ION, Signal transduction protein CBL-C | | Authors: | Takeshita, K, Tezuka, T, Isozaki, Y, Yamashita, E, Suzuki, M, Yamanashi, Y, Yamamoto, T, Nakagawa, A. | | Deposit date: | 2012-04-13 | | Release date: | 2013-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural flexibility regulates phosphopeptide-binding activity of the tyrosine kinase binding domain of Cbl-c.
J.Biochem., 152, 2012
|
|
