8J7M
 
 | | ion channel | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CHOLESTEROL, ion channel,Voltage dependent ion channel,Green fluorescent protein (Fragment),Voltage dependent ion channel,Green fluorescent protein (Fragment),Voltage dependent ion channel,Green fluorescent protein (Fragment) | | Authors: | Chen, H.W, Jiang, D. | | Deposit date: | 2023-04-27 | | Release date: | 2024-05-15 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural mechanism of voltage-gated sodium channel slow inactivation.
Nat Commun, 15, 2024
|
|
6PWS
 
 | |
6PWT
 
 | |
6O94
 
 | | Structure of the IRAK4 kinase domain with compound 17 | | Descriptor: | CALCIUM ION, Interleukin-1 receptor-associated kinase 4, N-{5-[4-(hydroxymethyl)piperidin-1-yl]-1-methyl-2-(morpholin-4-yl)-1H-benzimidazol-6-yl}pyrazolo[1,5-a]pyrimidine-3-carboxamide | | Authors: | Yu, C, Drobnick, J, Bryan, M.C, Kiefer, J, Lupardus, P.J. | | Deposit date: | 2019-03-13 | | Release date: | 2019-05-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Development of Potent and Selective Pyrazolopyrimidine IRAK4 Inhibitors.
J.Med.Chem., 62, 2019
|
|
6PWR
 
 | |
6O95
 
 | | Structure of the IRAK4 kinase domain with compound 41 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, N-[(2R)-2-(hydroxymethyl)-2-methyl-6-(morpholin-4-yl)-2,3-dihydro-1-benzofuran-5-yl]pyrazolo[1,5-a]pyrimidine-3-carboxamide, SULFATE ION | | Authors: | Yu, C, Drobnick, J, Bryan, M.C, Kiefer, J, Lupardus, P.J. | | Deposit date: | 2019-03-13 | | Release date: | 2019-05-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Development of Potent and Selective Pyrazolopyrimidine IRAK4 Inhibitors.
J.Med.Chem., 62, 2019
|
|
8GZ2
 
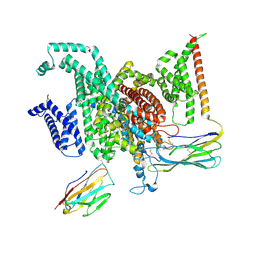 | | Cryo-EM structure of human NaV1.6/beta1/beta2-4,9-anhydro-tetrodotoxin | | Descriptor: | (1R,2S,3S,4R,5R,9S,11S,12S,14R)-7-amino-2,4,12-trihydroxy-2-(hydroxymethyl)-10,13,15-trioxa-6,8-diazapentacyclo[7.4.1.1~3,12~.0~5,11~.0~5,14~]pentadec-7-en-8-ium (non-preferred name), 2-acetamido-2-deoxy-beta-D-glucopyranose, Sodium channel protein type 8 subunit alpha, ... | | Authors: | Li, Y, Jiang, D. | | Deposit date: | 2022-09-24 | | Release date: | 2023-03-08 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of human Na V 1.6 channel reveals Na + selectivity and pore blockade by 4,9-anhydro-tetrodotoxin.
Nat Commun, 14, 2023
|
|
8GZ1
 
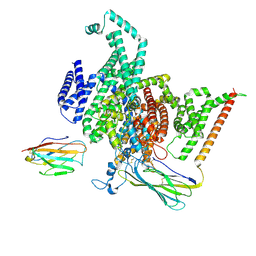 | | Cryo-EM structure of human NaV1.6/beta1/beta2,apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, Sodium channel protein type 8 subunit alpha, ... | | Authors: | Li, Y, Jiang, D. | | Deposit date: | 2022-09-24 | | Release date: | 2023-03-08 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of human Na V 1.6 channel reveals Na + selectivity and pore blockade by 4,9-anhydro-tetrodotoxin.
Nat Commun, 14, 2023
|
|
7CR5
 
 | |
8HKQ
 
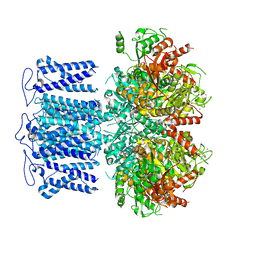 | | ion channel | | Descriptor: | POTASSIUM ION, Potassium channel subfamily T member 1, SODIUM ION, ... | | Authors: | Jiang, D.H, Zhang, J.T. | | Deposit date: | 2022-11-27 | | Release date: | 2023-08-16 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of human Slo2.2 channel gating and modulation.
Cell Rep, 42, 2023
|
|
8HKK
 
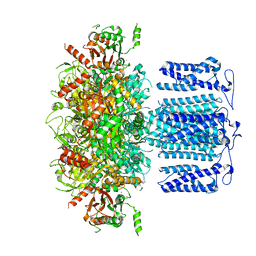 | | ion channel | | Descriptor: | POTASSIUM ION, Potassium channel subfamily T member 1, SODIUM ION, ... | | Authors: | Jiang, D.H, Zhang, J.T. | | Deposit date: | 2022-11-27 | | Release date: | 2023-08-16 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural basis of human Slo2.2 channel gating and modulation.
Cell Rep, 42, 2023
|
|
8HKF
 
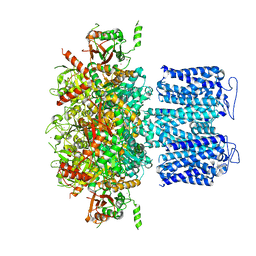 | | ion channel | | Descriptor: | POTASSIUM ION, Potassium channel subfamily T member 1, ZINC ION | | Authors: | Jiang, D.H, Zhang, Z.T. | | Deposit date: | 2022-11-25 | | Release date: | 2023-08-16 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural basis of human Slo2.2 channel gating and modulation.
Cell Rep, 42, 2023
|
|
8HKM
 
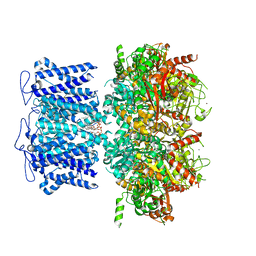 | | ion channel | | Descriptor: | POTASSIUM ION, Potassium channel subfamily T member 1, ZINC ION, ... | | Authors: | Jiang, D.H, Zhang, J.T. | | Deposit date: | 2022-11-27 | | Release date: | 2023-08-16 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis of human Slo2.2 channel gating and modulation.
Cell Rep, 42, 2023
|
|
8HK6
 
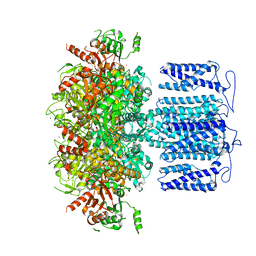 | | potassium channel | | Descriptor: | POTASSIUM ION, Potassium channel subfamily T member 1, ZINC ION | | Authors: | Jiang, D.H, Zhang, J.T. | | Deposit date: | 2022-11-25 | | Release date: | 2023-08-16 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Structural basis of human Slo2.2 channel gating and modulation.
Cell Rep, 42, 2023
|
|
8HIR
 
 | | potassium channels | | Descriptor: | POTASSIUM ION, Potassium channel subfamily T member 1, SODIUM ION, ... | | Authors: | Jiang, D.H, Zhang, J.T. | | Deposit date: | 2022-11-21 | | Release date: | 2023-08-16 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural basis of human Slo2.2 channel gating and modulation.
Cell Rep, 42, 2023
|
|
9IS8
 
 | | Cryo-EM structure of AKT1-AtKC1(G315D) | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, POTASSIUM ION, Potassium channel AKT1, ... | | Authors: | Dongliang, L, Zijie, Z, Yannan, Q, Yuyue, T, Huaizong, S. | | Deposit date: | 2024-07-17 | | Release date: | 2025-05-21 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | AtKC1 inhibits AKT1 activation via its amino-terminal inhibitory domain.
Plant Commun., 6, 2025
|
|
7E3O
 
 | |
7CI3
 
 | |
4KXF
 
 | |
8TUB
 
 | | HLA B7:02 with HPNGYKSLSTL | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Littler, D.R, Rossjohn, J, Chaurasia, P, Petersen, J. | | Deposit date: | 2023-08-16 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CD8 + T-cell responses towards conserved influenza B virus epitopes across anatomical sites and age.
Nat Commun, 15, 2024
|
|
8TUH
 
 | | HLA B7:02 with RPIIRPATL | | Descriptor: | 1,2-ETHANEDIOL, ARG-PRO-ILE-ILE-ARG-PRO-ALA-THR-LEU, Beta-2-microglobulin, ... | | Authors: | Littler, D.R, Rossjohn, J, Chaurasia, P, Petersen, J. | | Deposit date: | 2023-08-16 | | Release date: | 2024-03-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.200106 Å) | | Cite: | CD8 + T-cell responses towards conserved influenza B virus epitopes across anatomical sites and age.
Nat Commun, 15, 2024
|
|
8URF
 
 | |
7WLK
 
 | | CryoEM structure of human low-voltage activated T-type calcium channel Cav3.3 in complex with Otilonium Bromide(OB) | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-[diethyl(methyl)-$l^{4}-azanyl]ethyl 4-[(2-octoxyphenyl)carbonylamino]benzoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | He, L, Yu, Z, Dong, Y, Chen, Q, Zhao, Y. | | Deposit date: | 2022-01-13 | | Release date: | 2022-05-04 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure, gating, and pharmacology of human Ca V 3.3 channel.
Nat Commun, 13, 2022
|
|
7WLI
 
 | | CryoEM structure of human low-voltage activated T-type calcium channel CaV3.3 (apo) | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | He, L, Yu, Z, Dong, Y, Chen, Q, Zhao, Y. | | Deposit date: | 2022-01-13 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure, gating, and pharmacology of human Ca V 3.3 channel.
Nat Commun, 13, 2022
|
|
7WLL
 
 | | CryoEM structure of human low-voltage activated T-type calcium channel Cav3.3 in complex with pimozide(PMZ) | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[1-[4,4-bis(4-fluorophenyl)butyl]piperidin-4-yl]-1~{H}-benzimidazol-2-one, ... | | Authors: | He, L, Yu, Z, Dong, Y, Chen, Q, Zhao, Y. | | Deposit date: | 2022-01-13 | | Release date: | 2022-05-04 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure, gating, and pharmacology of human Ca V 3.3 channel.
Nat Commun, 13, 2022
|
|
