7SF7
 
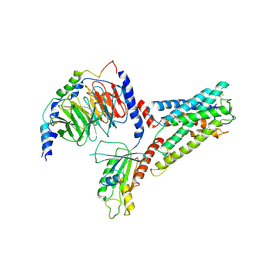 | | LPHN3 (ADGRL3) 7TM domain bound to tethered agonist in complex with G protein heterotrimer | | Descriptor: | G protein subunit 13 (Gi2-mini-G13 chimera), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Barros-Alvarez, X, Panova, O, Skiniotis, G. | | Deposit date: | 2021-10-03 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The tethered peptide activation mechanism of adhesion GPCRs.
Nature, 604, 2022
|
|
7T5Q
 
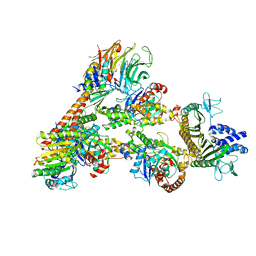 | | Cryo-EM Structure of a Transition State of Arp2/3 Complex Activation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Rebowski, G, van Eeuwen, T, Boczkowska, M, Dominguez, R. | | Deposit date: | 2021-12-13 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Transition State of Arp2/3 Complex Activation by Actin-Bound Dimeric Nucleation-Promoting Factor.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7SLX
 
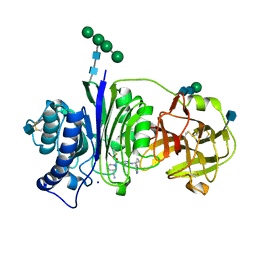 | | Vanin-1 complexed with Compound 11 | | Descriptor: | (3R)-1-(2-{[1-(pyrimidin-5-yl)cyclopropyl]amino}pyrimidine-5-carbonyl)piperidine-3-carbonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, Pantetheinase, ... | | Authors: | Vajdos, F.F. | | Deposit date: | 2021-10-25 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of a Series of Pyrimidine Carboxamides as Inhibitors of Vanin-1.
J.Med.Chem., 65, 2022
|
|
7SLV
 
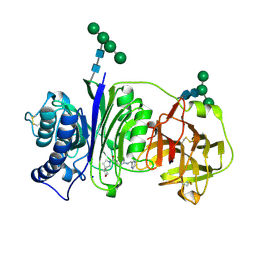 | | Vanin-1 complexed with Compound 3 | | Descriptor: | (8-oxa-2-azaspiro[4.5]decan-2-yl)(2-{[(pyrazin-2-yl)methyl]amino}pyrimidin-5-yl)methanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, Pantetheinase, ... | | Authors: | Vajdos, F.F. | | Deposit date: | 2021-10-25 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Discovery of a Series of Pyrimidine Carboxamides as Inhibitors of Vanin-1.
J.Med.Chem., 65, 2022
|
|
7SLY
 
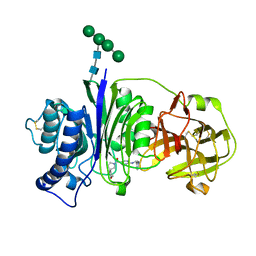 | | Vanin-1 complexed with Compound 27 | | Descriptor: | (8-oxa-2-azaspiro[4.5]decan-2-yl)(2-{[(1S)-1-(pyrazin-2-yl)ethyl]amino}pyrimidin-5-yl)methanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, Pantetheinase, ... | | Authors: | Vajdos, F.F. | | Deposit date: | 2021-10-25 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Discovery of a Series of Pyrimidine Carboxamides as Inhibitors of Vanin-1.
J.Med.Chem., 65, 2022
|
|
5ITH
 
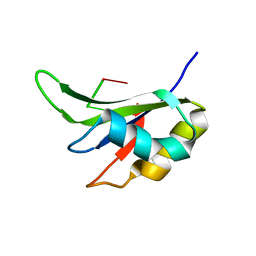 | |
5HL3
 
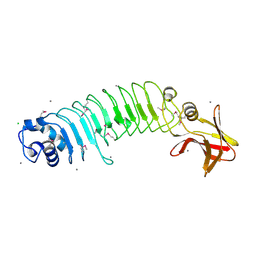 | | Crystal structure of Listeria monocytogenes InlP | | Descriptor: | CALCIUM ION, CHLORIDE ION, Lmo2470 protein | | Authors: | Nocadello, S, Light, S.H, Minasov, G, Kiryukhina, O, Kwon, K, Faralla, C, Bakardjiev, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-14 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Listeria monocytogenes InlP interacts with afadin and facilitates basement membrane crossing.
Plos Pathog., 14, 2018
|
|
1B10
 
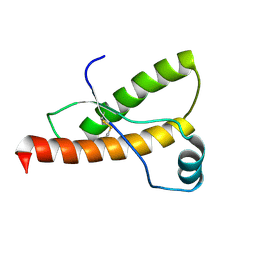 | | SOLUTION NMR STRUCTURE OF RECOMBINANT SYRIAN HAMSTER PRION PROTEIN RPRP(90-231) , 25 STRUCTURES | | Descriptor: | PROTEIN (PRION PROTEIN) | | Authors: | James, T.L, Liu, H, Ulyanov, N.B, Farr-Jones, S. | | Deposit date: | 1998-11-25 | | Release date: | 1998-12-02 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a 142-residue recombinant prion protein corresponding to the infectious fragment of the scrapie isoform.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
5IU5
 
 | | Lambda-[Ru(TAP)2(dppz)]2+ bound to d(TCGGCICCGA)2 | | Descriptor: | BARIUM ION, DNA (5'-D(*TP*CP*GP*GP*CP*IP*CP*CP*GP*A)-3'), Ru(tap)2(dppz) complex | | Authors: | Hall, J.P, Cardin, C.J. | | Deposit date: | 2016-03-17 | | Release date: | 2017-06-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unexpected enhancement of sensitised photo-oxidation by a Ru(II) complex on replacement of guanine with inosine in DNA
To Be Published
|
|
5UIS
 
 | | Crystal structure of IRAK4 in complex with compound 12 | | Descriptor: | 4-{[(3R)-piperidin-3-yl]oxy}-6-[(propan-2-yl)oxy]quinoline-7-carboxamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Han, S, Chang, J.S. | | Deposit date: | 2017-01-14 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of Clinical Candidate 1-{[(2S,3S,4S)-3-Ethyl-4-fluoro-5-oxopyrrolidin-2-yl]methoxy}-7-methoxyisoquinoline-6-carboxamide (PF-06650833), a Potent, Selective Inhibitor of Interleukin-1 Receptor Associated Kinase 4 (IRAK4), by Fragment-Based Drug Design.
J. Med. Chem., 60, 2017
|
|
5UIQ
 
 | | Crystal structure of IRAK4 in complex with compound 9 | | Descriptor: | 2-[(propan-2-yl)oxy]benzamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Han, S, Chang, J.S. | | Deposit date: | 2017-01-14 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Discovery of Clinical Candidate 1-{[(2S,3S,4S)-3-Ethyl-4-fluoro-5-oxopyrrolidin-2-yl]methoxy}-7-methoxyisoquinoline-6-carboxamide (PF-06650833), a Potent, Selective Inhibitor of Interleukin-1 Receptor Associated Kinase 4 (IRAK4), by Fragment-Based Drug Design.
J. Med. Chem., 60, 2017
|
|
5UIR
 
 | | Crystal structure of IRAK4 in complex with compound 11 | | Descriptor: | 5-(4-cyanophenyl)-3-[(propan-2-yl)oxy]naphthalene-2-carboxamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Han, S, Chang, J.S. | | Deposit date: | 2017-01-14 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Discovery of Clinical Candidate 1-{[(2S,3S,4S)-3-Ethyl-4-fluoro-5-oxopyrrolidin-2-yl]methoxy}-7-methoxyisoquinoline-6-carboxamide (PF-06650833), a Potent, Selective Inhibitor of Interleukin-1 Receptor Associated Kinase 4 (IRAK4), by Fragment-Based Drug Design.
J. Med. Chem., 60, 2017
|
|
5UIU
 
 | | Crystal structure of IRAK4 in complex with compound 30 | | Descriptor: | 1-{[(2S,3S,4S)-3-ethyl-4-fluoro-5-oxopyrrolidin-2-yl]methoxy}-7-methoxyisoquinoline-6-carboxamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Han, S, Chang, J.S. | | Deposit date: | 2017-01-14 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Discovery of Clinical Candidate 1-{[(2S,3S,4S)-3-Ethyl-4-fluoro-5-oxopyrrolidin-2-yl]methoxy}-7-methoxyisoquinoline-6-carboxamide (PF-06650833), a Potent, Selective Inhibitor of Interleukin-1 Receptor Associated Kinase 4 (IRAK4), by Fragment-Based Drug Design.
J. Med. Chem., 60, 2017
|
|
5UZU
 
 | | Immune evasion by a Staphylococcal Peroxidase Inhibitor that blocks myeloperoxidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | de Jong, N, Geisbrecht, B.V, van Strijp, J, Haas, P, Nijland, R, Ramyar, K, Fevre, C, Guerra, F, Voyich-Kane, J, van Kessel, C, Garcia, B. | | Deposit date: | 2017-02-27 | | Release date: | 2017-07-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Immune evasion by a staphylococcal inhibitor of myeloperoxidase.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5UIT
 
 | | Crystal structure of IRAK4 in complex with compound 14 | | Descriptor: | 1-{[(2S)-5-oxopyrrolidin-2-yl]methoxy}-7-[(propan-2-yl)oxy]isoquinoline-6-carboxamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Han, S, Chang, J.S. | | Deposit date: | 2017-01-14 | | Release date: | 2017-05-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Discovery of Clinical Candidate 1-{[(2S,3S,4S)-3-Ethyl-4-fluoro-5-oxopyrrolidin-2-yl]methoxy}-7-methoxyisoquinoline-6-carboxamide (PF-06650833), a Potent, Selective Inhibitor of Interleukin-1 Receptor Associated Kinase 4 (IRAK4), by Fragment-Based Drug Design.
J. Med. Chem., 60, 2017
|
|
8ORC
 
 | | Mus Musculus Acetylcholinesterase in complex with AL237 | | Descriptor: | 1-[2-(dimethylamino)ethyl]-3-(2-methoxyphenyl)thiourea, 2,5,8,11,14,17,20,23-OCTAOXAPENTACOSAN-25-OL, 2-(2-METHOXYETHOXY)ETHANOL, ... | | Authors: | Ekstrom, F.E, Linusson, A. | | Deposit date: | 2023-04-13 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Enzyme Dynamics Determine the Potency and Selectivity of Inhibitors Targeting Disease-Transmitting Mosquitoes.
Acs Infect Dis., 10, 2024
|
|
7Y1F
 
 | | Cryo-EM structure of human k-opioid receptor-Gi complex | | Descriptor: | Dynorphin, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Chen, B.O, Xu, F.E. | | Deposit date: | 2022-06-08 | | Release date: | 2023-05-24 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of human kappa-opioid receptor-Gi complex bound to an endogenous agonist dynorphin A.
Protein Cell, 14, 2023
|
|
1ZTQ
 
 | | Crystal structure of the catalytic domain of MMP-13 complexed with WAY-033 | | Descriptor: | CALCIUM ION, Collagenase 3, N-({4'-[(1-BENZOFURAN-2-YLCARBONYL)AMINO]-1,1'-BIPHENYL-4-YL}SULFONYL)-L-VALINE, ... | | Authors: | Wu, J, Rush III, T.S, Hotchandani, R, Du, X, Geck, M, Collins, E, Xu, Z.B, Skotnicki, J, Levin, J.I, Lovering, F. | | Deposit date: | 2005-05-27 | | Release date: | 2006-05-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of potent and selective MMP-13 inhibitors
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1NNQ
 
 | | rubrerythrin from Pyrococcus furiosus Pfu-1210814 | | Descriptor: | Rubrerythrin, ZINC ION | | Authors: | Liu, Z.-J, Tempel, W, Schubot, F.D, Shah, A, Arendall III, W.B, Rose, J.P, Richardson, D.C, Richardson, J.S, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-01-14 | | Release date: | 2004-03-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural genomics of Pyrococcus furiosus: X-ray crystallography reveals 3D domain swapping in rubrerythrin
Proteins, 57, 2004
|
|
4LKQ
 
 | | Crystal structure of PDE10A2 with fragment ZT017 | | Descriptor: | 4-amino-1,7-dihydro-6H-pyrazolo[3,4-d]pyrimidine-6-thione, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-07-08 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography.
J Biomol Screen, 19, 2014
|
|
4LLP
 
 | | Crystal structure of PDE10A2 with fragment ZT401 | | Descriptor: | 4-amino-2-methylphenol, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography.
J Biomol Screen, 19, 2014
|
|
4LM4
 
 | | Crystal structure of PDE10A2 with fragment ZT902 | | Descriptor: | NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A, quinazolin-4(1H)-one | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-07-10 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography.
J Biomol Screen, 19, 2014
|
|
4LM1
 
 | | Crystal structure of PDE10A2 with fragment ZT450 | | Descriptor: | 5-nitroquinoline, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography.
J Biomol Screen, 19, 2014
|
|
4LLX
 
 | | Crystal structure of PDE10A2 with fragment ZT434 | | Descriptor: | 4,6-dimethylpyrimidin-2-amine, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography.
J Biomol Screen, 19, 2014
|
|
4LM3
 
 | | Crystal structure of PDE10A2 with fragment ZT464 | | Descriptor: | 1,2-ETHANEDIOL, 1-(2,3-dihydro-1,4-benzodioxin-6-yl)ethanone, NICKEL (II) ION, ... | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography.
J Biomol Screen, 19, 2014
|
|
