4TKP
 
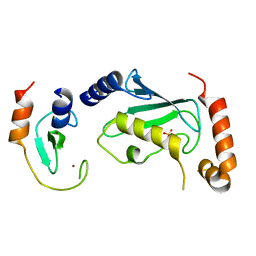 | | Complex of Ubc13 with the RING domain of the TRIM5alpha retroviral restriction factor | | Descriptor: | SULFATE ION, Tripartite motif-containing protein 5, Ubiquitin-conjugating enzyme E2 N, ... | | Authors: | Johnson, R, Taylor, A.B, Hart, P.J, Ivanov, D.N. | | Deposit date: | 2014-05-27 | | Release date: | 2015-07-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | RING Dimerization Links Higher-Order Assembly of TRIM5 alpha to Synthesis of K63-Linked Polyubiquitin.
Cell Rep, 12, 2015
|
|
4TLV
 
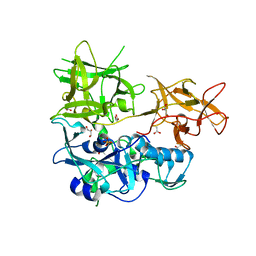 | | CARDS TOXIN, NICKED | | Descriptor: | ACETATE ION, ADP-ribosylating toxin CARDS, GLYCEROL, ... | | Authors: | Taylor, A.B, Pakhomova, O.N, Hart, P.J. | | Deposit date: | 2014-05-30 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of CARDS toxin, a unique ADP-ribosylating and vacuolating cytotoxin from Mycoplasma pneumoniae.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4RNX
 
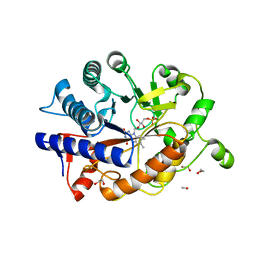 | | K154 Circular Permutation of Old Yellow Enzyme | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, NADPH dehydrogenase 1 | | Authors: | Horton, J.R, Daugherty, A.B, Cheng, X, Lutz, S. | | Deposit date: | 2014-10-26 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | STRUCTURAL AND FUNCTIONAL CONSEQUENCES OF CIRCULAR PERMUTATION ON THE ACTIVE SITE OF OLD YELLOW ENZYME.
ACS Catal, 5, 2015
|
|
4RP7
 
 | |
4RNU
 
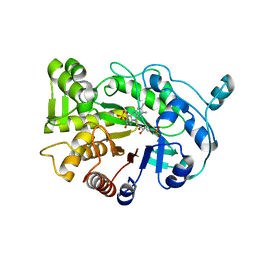 | | G303 Circular Permutation of Old Yellow Enzyme | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADPH dehydrogenase 1, PHOSPHATE ION | | Authors: | Horton, J.R, Daugherty, A.B, Cheng, X, Lutz, S. | | Deposit date: | 2014-10-26 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.677 Å) | | Cite: | STRUCTURAL AND FUNCTIONAL CONSEQUENCES OF CIRCULAR PERMUTATION ON THE ACTIVE SITE OF OLD YELLOW ENZYME.
ACS Catal, 5, 2015
|
|
4RP6
 
 | |
4TXW
 
 | | Crystal structure of CBM32-4 from the Clostridium perfringens NagH | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Hyaluronoglucosaminidase | | Authors: | Grondin, J.M, Ficko-Blean, E, Boraston, A.B, Smith, S.P. | | Deposit date: | 2014-07-07 | | Release date: | 2015-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Solution Structure and Dynamics of Full-length GH84A, a multimodular B-N-acetylglucosaminidase from Clostridium perfringens
To Be Published
|
|
4RNV
 
 | | G303 Circular Permutation of Old Yellow Enzyme with the Inhibitor p-Hydroxybenzaldehyde | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADPH dehydrogenase 1, P-HYDROXYBENZALDEHYDE | | Authors: | Horton, J.R, Daugherty, A.B, Cheng, X, Lutz, S. | | Deposit date: | 2014-10-26 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.473 Å) | | Cite: | STRUCTURAL AND FUNCTIONAL CONSEQUENCES OF CIRCULAR PERMUTATION ON THE ACTIVE SITE OF OLD YELLOW ENZYME.
ACS Catal, 5, 2015
|
|
4UAP
 
 | | X-ray structure of GH31 CBM32-2 bound to GalNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Grondin, J.M, Abe, K, Boraston, A.B, Smith, S.P. | | Deposit date: | 2014-08-11 | | Release date: | 2015-10-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Diverse modes of galacto-specific carbohydrate recognition by a family 31 glycoside hydrolase from Clostridium perfringens.
PLoS ONE, 12, 2017
|
|
3IK5
 
 | |
3IOZ
 
 | |
3KBL
 
 | | Crystal structure of the GLD-1 homodimerization domain from Caenorhabditis elegans N169A mutant at 2.28 A resolution | | Descriptor: | Female germline-specific tumor suppressor gld-1 | | Authors: | Beuck, C, Szymczyna, B.R, Kerkow, D.E, Carmel, A.B, Columbus, L, Stanfield, R.L, Williamson, J.R. | | Deposit date: | 2009-10-20 | | Release date: | 2010-03-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure of the GLD-1 homodimerization domain: insights into STAR protein-mediated translational regulation.
Structure, 18, 2010
|
|
3KBE
 
 | | Metal-free C. elegans Cu,Zn Superoxide Dismutase | | Descriptor: | Superoxide dismutase [Cu-Zn] | | Authors: | Pakhomova, O.N, Taylor, A.B, Schuermann, J.P, Culotta, V.L, Hart, P.J. | | Deposit date: | 2009-10-20 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | X-ray Crystal Structure of C. elegans Cu,Zn Superoxide Dismutase
To be Published
|
|
3KN7
 
 | |
3KBF
 
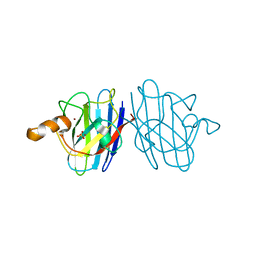 | | C. elegans Cu,Zn Superoxide Dismutase | | Descriptor: | COPPER (II) ION, SULFATE ION, Superoxide dismutase [Cu-Zn], ... | | Authors: | Pakhomova, O.N, Taylor, A.B, Schuermann, J.P, Culotta, V.L, Hart, P.J. | | Deposit date: | 2009-10-20 | | Release date: | 2010-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | X-ray Crystal Structure of C. elegans Cu,Zn Superoxide Dismutase
To be Published
|
|
3KLY
 
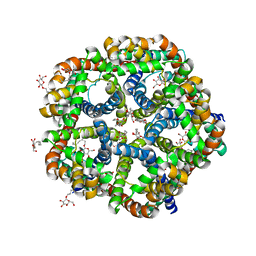 | |
3JBW
 
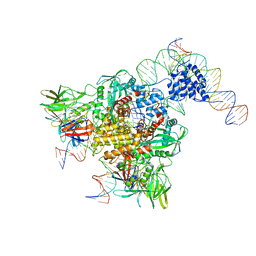 | | Cryo-electron microscopy structure of RAG Paired Complex (with NBD, no symmetry) | | Descriptor: | 12-RSS signal end forward strand, 5'-D(P*GP*AP*TP*CP*TP*GP*GP*CP*CP*TP*GP*TP*CP*TP*TP*A)-3', Nicked 12-RSS intermediate reverse strand, ... | | Authors: | Ru, H, Chambers, M.G, Fu, T.-M, Tong, A.B, Liao, M, Wu, H. | | Deposit date: | 2015-10-21 | | Release date: | 2015-12-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Molecular Mechanism of V(D)J Recombination from Synaptic RAG1-RAG2 Complex Structures.
Cell(Cambridge,Mass.), 163, 2015
|
|
3K6T
 
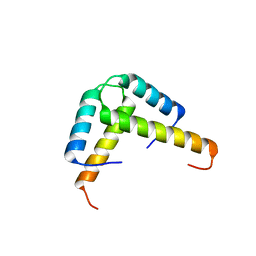 | | Crystal structure of the GLD-1 homodimerization domain from Caenorhabditis elegans at 2.04 A resolution | | Descriptor: | Female germline-specific tumor suppressor gld-1 | | Authors: | Beuck, C, Szymczyna, B.R, Kerkow, D.E, Carmel, A.B, Columbus, L, Stanfield, R.L, Williamson, J.R. | | Deposit date: | 2009-10-09 | | Release date: | 2010-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of the GLD-1 homodimerization domain: insights into STAR protein-mediated translational regulation.
Structure, 18, 2010
|
|
3K91
 
 | | Polysulfane Bridge in Cu-Zn Superoxide Dismutase | | Descriptor: | PENTASULFIDE-SULFUR, Superoxide dismutase [Cu-Zn] | | Authors: | You, Z, Cao, X, Taylor, A.B, Hart, P.J, Levine, R.L. | | Deposit date: | 2009-10-15 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Characterization of a covalent polysulfane bridge in copper-zinc superoxide dismutase .
Biochemistry, 49, 2010
|
|
3JBL
 
 | | Cryo-EM Structure of the Activated NAIP2/NLRC4 Inflammasome Reveals Nucleated Polymerization | | Descriptor: | NLR family CARD domain-containing protein 4 | | Authors: | Zhang, L, Chen, S, Ruan, J, Wu, J, Tong, A.B, Yin, Q, Li, Y, David, L, Lu, A, Wang, W.L, Marks, C, Ouyang, Q, Zhang, X, Mao, Y, Wu, H. | | Deposit date: | 2015-09-05 | | Release date: | 2015-10-21 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization.
Science, 350, 2015
|
|
3JBX
 
 | | Cryo-electron microscopy structure of RAG Signal End Complex (C2 symmetry) | | Descriptor: | 5'-D(*CP*AP*CP*AP*GP*TP*GP*CP*TP*AP*CP*AP*GP*AP*C)-3', 5'-D(*GP*CP*GP*AP*TP*GP*GP*TP*TP*AP*AP*CP*CP*A)-3', 5'-D(P*GP*TP*CP*TP*GP*TP*AP*GP*CP*AP*CP*TP*GP*TP*G)-3', ... | | Authors: | Ru, H, Chambers, M.G, Fu, T.-M, Tong, A.B, Liao, M, Wu, H. | | Deposit date: | 2015-10-22 | | Release date: | 2015-12-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular Mechanism of V(D)J Recombination from Synaptic RAG1-RAG2 Complex Structures.
Cell(Cambridge,Mass.), 163, 2015
|
|
3KM2
 
 | |
3IXS
 
 | | Ring1B C-terminal domain/RYBP C-terminal domain Complex | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, E3 ubiquitin-protein ligase RING2, ... | | Authors: | Wang, R, Taylor, A.B, Kim, C.A. | | Deposit date: | 2009-09-04 | | Release date: | 2010-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Polycomb Group Targeting through Different Binding Partners of RING1B C-Terminal Domain.
Structure, 18, 2010
|
|
3J5M
 
 | | Cryo-EM structure of the BG505 SOSIP.664 HIV-1 Env trimer with 3 PGV04 Fabs | | Descriptor: | BG505 SOSIP gp120, BG505 SOSIP gp41, PGV04 heavy chain, ... | | Authors: | Lyumkis, D, Julien, J.-P, Wilson, I.A, Ward, A.B. | | Deposit date: | 2013-10-26 | | Release date: | 2013-11-13 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer.
Science, 342, 2013
|
|
1GM2
 
 | |
