4LA3
 
 | |
6K9M
 
 | | Human LXR-beta in complex with an agonist | | Descriptor: | Oxysterols receptor LXR-beta, ~{tert}-butyl (2'~{S},3~{S})-2-oxidanylidene-2'-propan-2-yl-spiro[1~{H}-indole-3,3'-pyrrolidine]-1'-carboxylate | | Authors: | Zhang, Z, Zhou, H. | | Deposit date: | 2019-06-16 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of novel liver X receptor inverse agonists as lipogenesis inhibitors.
Eur.J.Med.Chem., 206, 2020
|
|
5TDK
 
 | | RNA decamer duplex with eight 2'-5'-linkages | | Descriptor: | RNA (5'-R(*CP*CP*GP*GP*CP*GP*CP*CP*GP*G)-3'), STRONTIUM ION | | Authors: | Luo, Z, Sheng, J. | | Deposit date: | 2016-09-19 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural insights into RNA duplexes with multiple 2 -5 -linkages.
Nucleic Acids Res., 45, 2017
|
|
2F9Z
 
 | | Complex between the chemotaxis deamidase CheD and the chemotaxis phosphatase CheC from Thermotoga maritima | | Descriptor: | PROTEIN (chemotaxis methylation protein), chemotaxis protein CheC | | Authors: | Chao, X, Park, S.Y, Bilwes, A.M, Crane, B.R. | | Deposit date: | 2005-12-06 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A receptor-modifying deamidase in complex with a signaling phosphatase reveals reciprocal regulation.
Cell(Cambridge,Mass.), 124, 2006
|
|
5TDJ
 
 | | RNA decamer duplex with four 2'-5'-linkages | | Descriptor: | RNA (5'-R(*CP*CP*GP*GP*CP*GP*CP*CP*GP*G)-3'), STRONTIUM ION | | Authors: | Luo, Z, Sheng, J. | | Deposit date: | 2016-09-19 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into RNA duplexes with multiple 2 -5 -linkages.
Nucleic Acids Res., 45, 2017
|
|
4I6P
 
 | | Crystal structure of Par3-NTD domain | | Descriptor: | Partitioning defective 3 homolog | | Authors: | Wang, W, Gao, F, Gong, W, Sun, F, Feng, W. | | Deposit date: | 2012-11-29 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into the intrinsic self-assembly of par-3 N-terminal domain.
Structure, 21, 2013
|
|
2KHP
 
 | |
8UR5
 
 | |
1GGQ
 
 | |
8UR7
 
 | |
4LT4
 
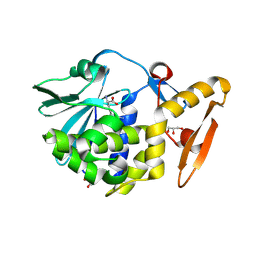 | | Crystal structure of arginine inhibited Ribosome inactivating protein from Momordica balsamina at 1.69 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ARGININE, GLYCEROL, ... | | Authors: | Yamini, S, Pandey, S, Bhushan, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-07-23 | | Release date: | 2013-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of arginine inhibited Ribosome inactivating protein from Momordica balsamina at 1.69 A resolution
To be Published
|
|
5K5E
 
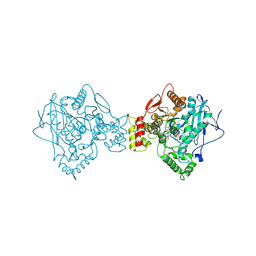 | | Discovery and Structure-Activity Relationships of a Highly Selective Butyrylcholinesterase Inhibitor by Structure-Based Virtual Screening | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | De la Mora, E, Dighe, S.N, Deora, G.S, Ross, B.P, Nachon, F, Brazzolotto, X. | | Deposit date: | 2016-05-23 | | Release date: | 2016-07-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and Structure-Activity Relationships of a Highly Selective Butyrylcholinesterase Inhibitor by Structure-Based Virtual Screening.
J.Med.Chem., 59, 2016
|
|
2K7X
 
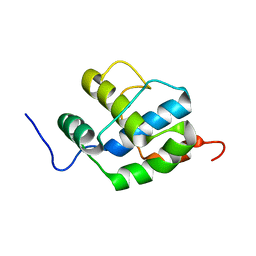 | |
1MAT
 
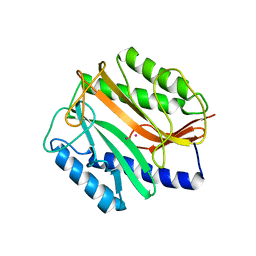 | |
1ML4
 
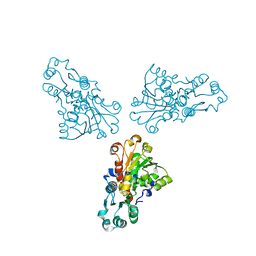 | |
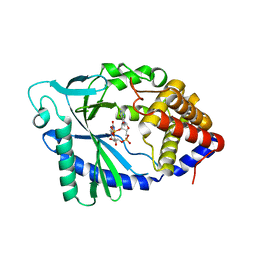
5VDR
 
 | |
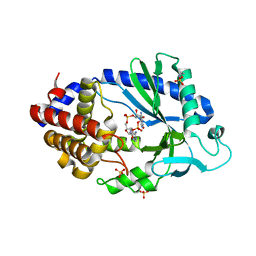
5VDP
 
 | |
5K98
 
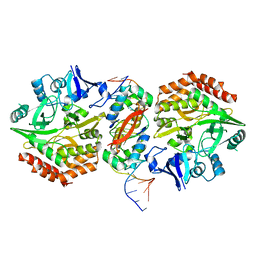 | | Structure of HipA-HipB-O2-O3 complex | | Descriptor: | Antitoxin HipB, DNA (5'-D(*CP*TP*AP*TP*CP*CP*CP*CP*TP*TP*AP*AP*GP*GP*GP*GP*AP*TP*AP*GP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*CP*TP*AP*TP*CP*CP*CP*CP*TP*TP*AP*AP*GP*GP*GP*GP*AP*TP*AP*G)-3'), ... | | Authors: | Schumacher, M. | | Deposit date: | 2016-05-31 | | Release date: | 2016-06-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.99 Å) | | Cite: | HipBA-promoter structures reveal the basis of heritable multidrug tolerance.
Nature, 524, 2015
|
|
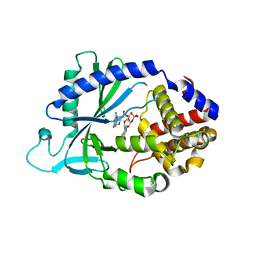
5VDQ
 
 | |
8V5P
 
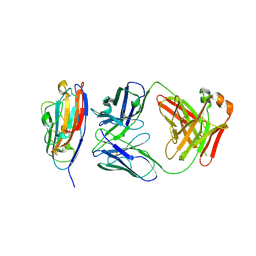 | |
8V5S
 
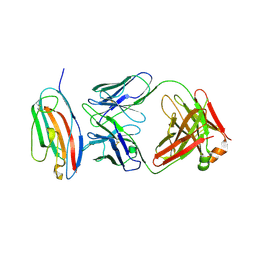 | |
8V5L
 
 | | Structure of the Varicella Zoster Virus (VZV) gI binding domain of glycoprotein E (gE) in complex with human Fab 1A2 and 1E12 | | Descriptor: | Envelope glycoprotein E, Fab 1A2 Heavy Chain, Fab 1A2 Light Chain, ... | | Authors: | Seraj, N, Holzapfel, G, Harshbarger, W. | | Deposit date: | 2023-11-30 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structures of the Varicella Zoster Virus Glycoprotein E and Epitope Mapping of Vaccine-Elicited Antibodies
Vaccines (Basel), 12, 2024
|
|
3G5O
 
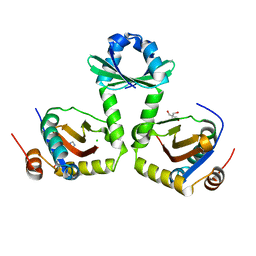 | | The crystal structure of the toxin-antitoxin complex RelBE2 (Rv2865-2866) from Mycobacterium tuberculosis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Miallau, L, Cascio, D, Eisenberg, D, TB Structural Genomics Consortium (TBSGC), Integrated Center for Structure and Function Innovation (ISFI) | | Deposit date: | 2009-02-05 | | Release date: | 2009-04-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Comparative proteomics identifies the cell-associated lethality of M. tuberculosis RelBE-like toxin-antitoxin complexes.
Structure, 21, 2013
|
|
5W6X
 
 | |
2RJZ
 
 | | Crystal structure of the type 4 fimbrial biogenesis protein PilO from Pseudomonas aeruginosa | | Descriptor: | PilO protein, SULFATE ION | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Chang, S, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-10-16 | | Release date: | 2007-11-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Periplasmic domains of Pseudomonas aeruginosa PilN and PilO form a stable heterodimeric complex.
J.Mol.Biol., 394, 2009
|
|
