6ITA
 
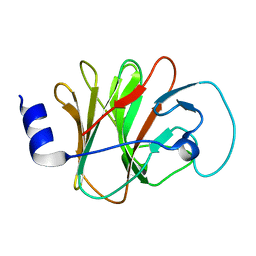 | | Crystal structure of intracellular B30.2 domain of BTN3A1 mutant | | Descriptor: | Butyrophilin subfamily 3 member A1 | | Authors: | Yang, Y.Y, Liu, W.D, Cai, N.N, Chen, C.C, Guo, R.T, Zhang, Y.H. | | Deposit date: | 2018-11-20 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | A Structural Change in Butyrophilin upon Phosphoantigen Binding Underlies Phosphoantigen-Mediated V gamma 9V delta 2 T Cell Activation.
Immunity, 50, 2019
|
|
1YGE
 
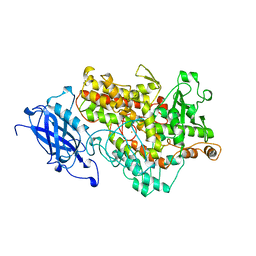 | | LIPOXYGENASE-1 (SOYBEAN) AT 100K | | Descriptor: | FE (III) ION, LIPOXYGENASE-1 | | Authors: | Minor, W, Steczko, J, Stec, B, Otwinowski, Z, Bolin, J.T, Walter, R, Axelrod, B. | | Deposit date: | 1996-06-04 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of soybean lipoxygenase L-1 at 1.4 A resolution.
Biochemistry, 35, 1996
|
|
6KLV
 
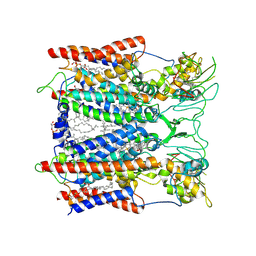 | | Hyperthermophilic respiratory Complex III | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 2-[(2~{E},6~{E},10~{Z},14~{Z},18~{Z},23~{R})-3,7,11,15,19,23,27-heptamethyloctacosa-2,6,10,14,18-pentaenyl]naphthalene-1,4-dione, ANTIMYCIN, ... | | Authors: | Fei, S, Hartmut, M, Yun, Z, Guohong, P, Guoliang, Z, Hui, Z, Shuangbo, Z, Xiaoyun, P, Yan, Z. | | Deposit date: | 2019-07-30 | | Release date: | 2020-05-13 | | Last modified: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A 3.3 angstrom -Resolution Structure of Hyperthermophilic Respiratory Complex III Reveals the Mechanism of Its Thermal Stability.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
2F5I
 
 | |
2YU2
 
 | | Crystal structure of hJHDM1A without a-ketoglutarate | | Descriptor: | FE (II) ION, JmjC domain-containing histone demethylation protein 1A | | Authors: | Han, Z. | | Deposit date: | 2007-04-05 | | Release date: | 2007-04-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for histone demethylation by JHDM1
To be Published
|
|
2YU1
 
 | | Crystal structure of hJHDM1A complexed with a-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, JmjC domain-containing histone demethylation protein 1A | | Authors: | Han, Z. | | Deposit date: | 2007-04-05 | | Release date: | 2007-04-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for histone demethylation by JHDM1
To be Published
|
|
8HOY
 
 | | Cryo-EM structure of monkeypox virus DNA replication holoenzyme F8, A22 and E4 complex without DNA at 2.76 angostram | | Descriptor: | DNA polymerase, DNA polymerase processivity factor component A20, E4R | | Authors: | Xu, Y, Wu, Y, Zhang, Y, Fan, R, Yang, Y, Li, D, Yang, B, Zhang, Z, Dong, C. | | Deposit date: | 2022-12-11 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structure of DNA replication machinery from human monkeypox virus
To Be Published
|
|
8HPA
 
 | | Monkeypox virus DNA replication holoenzyme F8, A22 and E4 complex in a DNA binding form | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*CP*CP*TP*TP*CP*CP*CP*CP*TP*AP*C)-3'), DNA (5'-D(P*AP*TP*GP*GP*TP*AP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*CP*G)-3'), DNA polymerase, ... | | Authors: | Xu, Y, Wu, Y, Zhang, Y, Fan, R, Yang, Y, Li, D, Yang, B, Zhang, Z, Dong, C. | | Deposit date: | 2022-12-12 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structure of DNA replication machinery from human monkeypox virus
To Be Published
|
|
8I3U
 
 | | Local CryoEM structure of the SARS-CoV-2 S6P in complex with 14B1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 14B1, Light chain of Fab 14B1, ... | | Authors: | Li, Z, Yu, F, Cao, S. | | Deposit date: | 2023-01-18 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Broadly neutralizing antibodies derived from the earliest COVID-19 convalescents protect mice from SARS-CoV-2 variants challenge.
Signal Transduct Target Ther, 8, 2023
|
|
8I3S
 
 | | Local CryoEM structure of the SARS-CoV-2 S6P in complex with 7B3 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain od Fab 7B3, Light chain of Fab 7B3, ... | | Authors: | Li, Z, Yu, F, Cao, S, ZHao, H. | | Deposit date: | 2023-01-17 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Broadly neutralizing antibodies derived from the earliest COVID-19 convalescents protect mice from SARS-CoV-2 variants challenge.
Signal Transduct Target Ther, 8, 2023
|
|
3A0E
 
 | | Crystal Structure of Polygonatum cyrtonema lectin (PCL) complexed with dimannoside | | Descriptor: | Mannose/sialic acid-binding lectin, SULFATE ION, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose | | Authors: | Ding, J, Wang, D.C. | | Deposit date: | 2009-03-16 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of a novel anti-HIV mannose-binding lectin from Polygonatum cyrtonema Hua with unique ligand-binding property and super-structure
J.Struct.Biol., 171, 2010
|
|
3A0C
 
 | |
3A0D
 
 | |
6MYN
 
 | | Crystal structure of murine NF-kappaB inducing kinase (NIK) bound to inhibitor R7 | | Descriptor: | (5s,7s)-9-fluoro-10-[(3R)-3-hydroxy-3-(5-methyl-1,2-oxazol-3-yl)but-1-yn-1-yl]-N~3~-methyl-6,7-dihydro-5H-5,7-methanoimidazo[2,1-a][2]benzazepine-2,3-dicarboxamide, Mitogen-activated protein kinase kinase kinase 14, SULFATE ION | | Authors: | Harris, S.F, Smith, M, Barker, J. | | Deposit date: | 2018-11-01 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.744 Å) | | Cite: | Structure Based Design of Potent Selective Inhibitors of Protein Kinase D1 (PKD1).
Acs Med.Chem.Lett., 10, 2019
|
|
7WP6
 
 | | Cryo-EM structure of SARS-CoV-2 recombinant spike protein STFK in complex with three neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 36H6 heavy chain, ... | | Authors: | Zheng, Q, Sun, H, Yuan, Q, Li, S, Xia, N. | | Deposit date: | 2022-01-23 | | Release date: | 2023-03-01 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Lineage-mosaic and mutation-patched spike proteins for broad-spectrum COVID-19 vaccine.
Cell Host Microbe, 30, 2022
|
|
7WP8
 
 | | Cryo-EM structure of SARS-CoV-2 recombinant spike protein STFK1628x in complex with three neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2B4 heavy chain, ... | | Authors: | Zheng, Q, Sun, H, Yuan, Q, Li, S, Xia, N. | | Deposit date: | 2022-01-23 | | Release date: | 2023-03-08 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Lineage-mosaic and mutation-patched spike proteins for broad-spectrum COVID-19 vaccine.
Cell Host Microbe, 30, 2022
|
|
8K46
 
 | | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses including all major Omicron strains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y, Gao, Y, Yao, H, Xu, W, Yang, H. | | Deposit date: | 2023-07-17 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses, including all major Omicron strains.
MedComm (2020), 4, 2023
|
|
8K45
 
 | | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses including all major Omicron strains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nb4 nanobody, ... | | Authors: | Lu, Y, Gao, Y, Yao, H, Xu, W, Yang, H. | | Deposit date: | 2023-07-17 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses, including all major Omicron strains.
MedComm (2020), 4, 2023
|
|
8K47
 
 | | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses including all major Omicron strains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y, Gao, Y, Yao, H, Xu, W, Yang, H. | | Deposit date: | 2023-07-17 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses, including all major Omicron strains.
MedComm (2020), 4, 2023
|
|
3AFK
 
 | |
3BJM
 
 | | Crystal structure of human DPP-IV in complex with (1S,3S, 5S)-2-[(2S)-2-AMINO-2-(3-HYDROXYTRICYCLO[3.3.1.13,7]DEC-1- YL)ACETYL]-2-AZABICYCLO[3.1.0]HEXANE-3-CARBONITRILE (CAS), (1S,3S,5S)-2-((2S)-2-AMINO-2-(3-HYDROXYADAMANTAN-1- YL)ACETYL)-2-AZABICYCLO[3.1.0]HEXANE-3-CARBONITRILE (IUPAC), OR BMS-477118 | | Descriptor: | (2~{S})-2-azanyl-1-[(1~{S},3~{S},5~{S})-3-(iminomethyl)-2-azabicyclo[3.1.0]hexan-2-yl]-2-[(5~{R},7~{S})-3-oxidanyl-1-ad amantyl]ethanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4 | | Authors: | Klei, H.E. | | Deposit date: | 2007-12-04 | | Release date: | 2008-04-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Involvement of DPP-IV catalytic residues in enzyme-saxagliptin complex formation.
Protein Sci., 17, 2008
|
|
6O8L
 
 | |
7XPZ
 
 | | Structure of Apo-hSLC19A1 | | Descriptor: | Reduced folate transporter | | Authors: | Zhang, Q.X, Zhang, X.Y, Zhu, Y.L, Sun, P.P, Gao, A, Zhang, L.G, Gao, P. | | Deposit date: | 2022-05-06 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Recognition of cyclic dinucleotides and folates by human SLC19A1.
Nature, 612, 2022
|
|
7XQ0
 
 | | Structure of hSLC19A1+3'3'-CDA | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, Reduced folate transporter | | Authors: | Zhang, Q.X, Zhang, X.Y, Zhu, Y.L, Sun, P.P, Gao, A, Zhang, L.G, Gao, P. | | Deposit date: | 2022-05-06 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Recognition of cyclic dinucleotides and folates by human SLC19A1.
Nature, 612, 2022
|
|
7XQ2
 
 | | Structure of hSLC19A1+2'3'-cGAMP | | Descriptor: | Reduced folate transporter, cGAMP | | Authors: | Zhang, Q.X, Zhang, X.Y, Zhu, Y.L, Sun, P.P, Gao, A, Zhang, L.G, Gao, P. | | Deposit date: | 2022-05-06 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Recognition of cyclic dinucleotides and folates by human SLC19A1.
Nature, 612, 2022
|
|
