6MIA
 
 | | Crystal structure of CTX-M-14 with compound 6 | | Descriptor: | 3-(1H-tetrazol-5-ylmethyl)-5,6,7,8-tetrahydro[1]benzothieno[2,3-d]pyrimidin-4(3H)-one, Beta-lactamase | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2018-09-19 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Active-Site Druggability of Carbapenemases and Broad-Spectrum Inhibitor Discovery.
Acs Infect Dis., 5, 2019
|
|
5XZ6
 
 | | Crystal Structure of Phosphofructokinase from Staphylococcus aureus in complex with adenylylimidodiphosphate, the ATP analogue | | Descriptor: | ATP-dependent 6-phosphofructokinase, GLYCEROL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Wang, C.L, Tian, T, Zang, J.Y. | | Deposit date: | 2017-07-11 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insights into the Regulation of Staphylococcus aureus Phosphofructokinase by Tetramer-Dimer Conversion.
Biochemistry, 57, 2018
|
|
6MD8
 
 | | Crystal structure of CTX-M-14 with compound 3 | | Descriptor: | 1-(2,4-dichlorophenyl)-4-(1H-tetrazol-5-yl)-1H-pyrazol-5-amine, Beta-lactamase CTX-M-14, DIMETHYL SULFOXIDE | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2018-09-04 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Active-Site Druggability of Carbapenemases and Broad-Spectrum Inhibitor Discovery.
Acs Infect Dis., 5, 2019
|
|
8IQF
 
 | | Cryo-EM structure of the dimeric human CAF1-H3-H4 complex | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, Z.Y, Xu, R.M. | | Deposit date: | 2023-03-16 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
8IQG
 
 | | Cryo-EM structure of the monomeric human CAF1-H3-H4 complex | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, Z.Y, Xu, R.M. | | Deposit date: | 2023-03-16 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
7X6Y
 
 | |
6NCL
 
 | |
7DRB
 
 | | Crystal structure of plant receptor like protein RXEG1 with xyloglucanase XEG1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cell 12A endoglucanase, ... | | Authors: | Sun, Y, Wang, Y, Zhang, X.X, Chen, Z.D, Xia, Y.Q, Sun, Y.J, Zhang, M.M, Xiao, Y, Han, Z.F, Wang, Y.C, Chai, J.J. | | Deposit date: | 2020-12-27 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Plant receptor-like protein activation by a microbial glycoside hydrolase.
Nature, 610, 2022
|
|
7DRC
 
 | | Cryo-EM structure of plant receptor like protein RXEG1 in complex with xyloglucanase XEG1 and BAK1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Brassinosteroid insensitive 1-associated receptor kinase 1, ... | | Authors: | Sun, Y, Wang, Y, Zhang, X.X, Chen, Z.D, Xia, Y.Q, Sun, Y.J, Zhang, M.M, Xiao, Y, Han, Z.F, Wang, Y.C, Chai, J.J. | | Deposit date: | 2020-12-27 | | Release date: | 2022-06-22 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Plant receptor-like protein activation by a microbial glycoside hydrolase.
Nature, 610, 2022
|
|
6MEY
 
 | | Crystal structure of KPC-2 with compound 9 | | Descriptor: | (2R)-2-phenyl-2-(phenylamino)-N-(1H-tetrazol-5-yl)acetamide, ACETATE ION, Carbapenem-hydrolyzing beta-lactamase KPC | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2018-09-07 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Active-Site Druggability of Carbapenemases and Broad-Spectrum Inhibitor Discovery.
Acs Infect Dis., 5, 2019
|
|
7X6Z
 
 | |
7X70
 
 | |
6MNP
 
 | | Crystal structure of KPC-2 with compound 6 | | Descriptor: | 3-(1H-tetrazol-5-ylmethyl)-5,6,7,8-tetrahydro[1]benzothieno[2,3-d]pyrimidin-4(3H)-one, Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2018-10-02 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Active-Site Druggability of Carbapenemases and Broad-Spectrum Inhibitor Discovery.
Acs Infect Dis., 5, 2019
|
|
6MLL
 
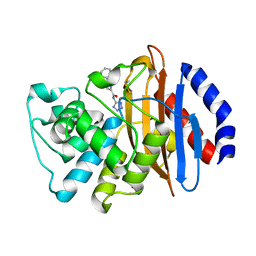 | | Crystal structure of KPC-2 with compound 7 | | Descriptor: | 1,5-diphenyl-N-(1H-tetrazol-5-yl)-1H-pyrazole-3-carboxamide, Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2018-09-27 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Active-Site Druggability of Carbapenemases and Broad-Spectrum Inhibitor Discovery.
Acs Infect Dis., 5, 2019
|
|
6M53
 
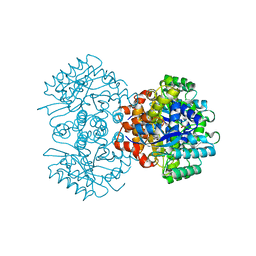 | | Crystal structure of 2, 3-dihydroxybenzoic acid decarboxylase from Fusarium oxysporum | | Descriptor: | 2,3-dihydroxybenzoate decarboxylase, GLYCEROL, ZINC ION | | Authors: | Song, M.K, Feng, J.H, Liu, W.D, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2020-03-09 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 2,3-Dihydroxybenzoic Acid Decarboxylase from Fusarium oxysporum: Crystal Structures and Substrate Recognition Mechanism.
Chembiochem, 21, 2020
|
|
6MDU
 
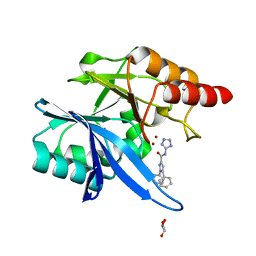 | | Crystal structure of NDM-1 with compound 7 | | Descriptor: | 1,5-diphenyl-N-(1H-tetrazol-5-yl)-1H-pyrazole-3-carboxamide, GLYCEROL, Metallo-beta-lactamase type 2, ... | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Active-Site Druggability of Carbapenemases and Broad-Spectrum Inhibitor Discovery.
Acs Infect Dis., 5, 2019
|
|
7XPQ
 
 | |
7XPP
 
 | |
7XPO
 
 | | Crystal Structure of UDP-Glc/GlcNAc 4-Epimerase with NAD/UDP-Glc | | Descriptor: | GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 4-epimerase, ... | | Authors: | Chen, Y.H, Wang, X.C, Zhang, C.R. | | Deposit date: | 2022-05-05 | | Release date: | 2023-05-17 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A maize epimerase modulates cell wall synthesis and glycosylation during stomatal morphogenesis.
Nat Commun, 14, 2023
|
|
8HL3
 
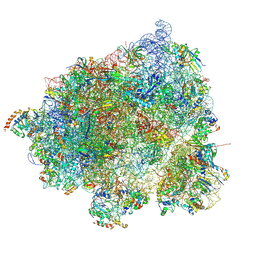 | |
8HL5
 
 | |
8HL4
 
 | |
7DBI
 
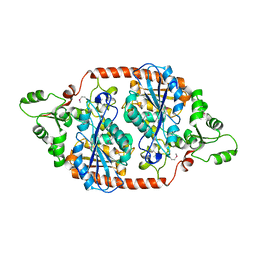 | | Crystal structure of the peroxisomal acyl-CoA hydrolase MpaH | | Descriptor: | acyl-CoA hydrolase MpaH' | | Authors: | Li, S.Y, You, C. | | Deposit date: | 2020-10-20 | | Release date: | 2021-04-28 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis for substrate specificity of the peroxisomal acyl-CoA hydrolase MpaH' involved in mycophenolic acid biosynthesis.
Febs J., 288, 2021
|
|
7DBL
 
 | | Acyl-CoA hydrolase MpaH' mutant S139A in complex with MPA | | Descriptor: | MYCOPHENOLIC ACID, acyl-CoA hydrolase MpaH' | | Authors: | Li, S.Y, You, C. | | Deposit date: | 2020-10-20 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural basis for substrate specificity of the peroxisomal acyl-CoA hydrolase MpaH' involved in mycophenolic acid biosynthesis.
Febs J., 288, 2021
|
|
8HQL
 
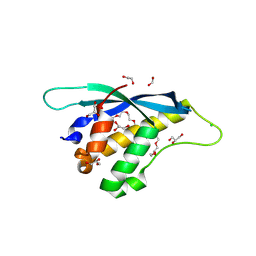 | | Crystal structure of mouse SNX25 PX domain | | Descriptor: | ACETIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Yu, Z, Xu, J, Liu, J. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-20 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Redox-modulated SNX25 as a novel regulator of GPCR-G protein signaling from endosomes.
Redox Biol, 75, 2024
|
|
