8JSW
 
 | | Human VMAT2 complex with serotonin | | Descriptor: | SEROTONIN, Synaptic vesicular amine transporter | | Authors: | Jiang, D.H, Wu, D. | | Deposit date: | 2023-06-20 | | Release date: | 2023-11-29 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Transport and inhibition mechanisms of human VMAT2.
Nature, 626, 2024
|
|
8JTC
 
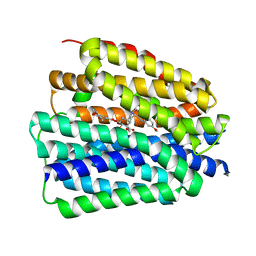 | | Human VMAT2 complex with reserpine | | Descriptor: | Synaptic vesicular amine transporter, reserpine | | Authors: | Jiang, D.H, Wu, D. | | Deposit date: | 2023-06-21 | | Release date: | 2023-11-29 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Transport and inhibition mechanisms of human VMAT2.
Nature, 626, 2024
|
|
8JT9
 
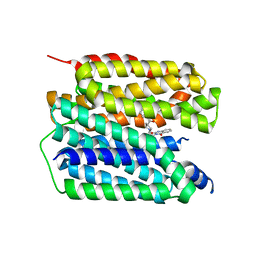 | | Human VMAT2 complex with ketanserin | | Descriptor: | 3-[2-[4-(4-fluorophenyl)carbonylpiperidin-1-yl]ethyl]-1~{H}-quinazoline-2,4-dione, Synaptic vesicular amine transporter | | Authors: | Jiang, D.H, Wu, D. | | Deposit date: | 2023-06-21 | | Release date: | 2023-11-29 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Transport and inhibition mechanisms of human VMAT2.
Nature, 626, 2024
|
|
8JTA
 
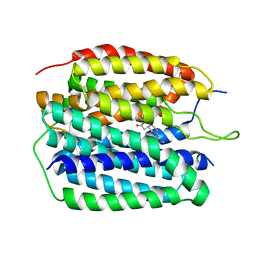 | | Human VMAT2 complex with tetrabenazine | | Descriptor: | Synaptic vesicular amine transporter, tetrabenazine | | Authors: | Jiang, D.H, Wu, D. | | Deposit date: | 2023-06-21 | | Release date: | 2023-11-29 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Transport and inhibition mechanisms of human VMAT2.
Nature, 626, 2024
|
|
5B25
 
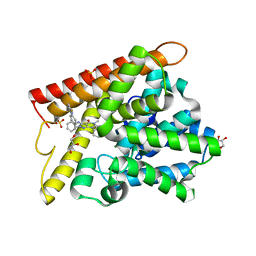 | | Crystal structure of human PDE1B with inhibitor 3 | | Descriptor: | (11R,15S)-4-{[4-(6-fluoropyridin-2-yl)phenyl]methyl}-8-methyl-5-(phenylamino)-1,3,4,8,10-pentaazatetracyclo[7.6.0.02,6.011,15]pentadeca-2,5,9-trien-7-one, Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B, GLYCEROL, ... | | Authors: | Ida, K, Lane, W, Snell, G, Sogabe, S. | | Deposit date: | 2016-01-07 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Potent and Selective Inhibitors of Phosphodiesterase 1 for the Treatment of Cognitive Impairment Associated with Neurodegenerative and Neuropsychiatric Diseases
J.Med.Chem., 59, 2016
|
|
5BKH
 
 | |
8JGR
 
 | |
8JGT
 
 | |
8JGX
 
 | |
8JGW
 
 | |
8JGU
 
 | |
8JGQ
 
 | |
8JGP
 
 | |
8JGO
 
 | |
7BZG
 
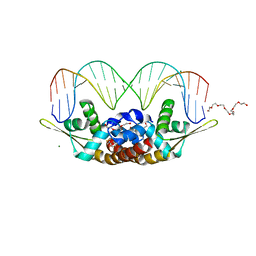 | | Structure of Bacillus subtilis HxlR, wild type in complex with formaldehyde and DNA | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*CP*AP*GP*TP*AP*TP*CP*CP*TP*CP*GP*AP*GP*GP*AP*TP*AP*CP*TP*G)-3'), ... | | Authors: | Zhu, R, Chen, P.R. | | Deposit date: | 2020-04-27 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Genetically encoded formaldehyde sensors inspired by a protein intra-helical crosslinking reaction.
Nat Commun, 12, 2021
|
|
7BZD
 
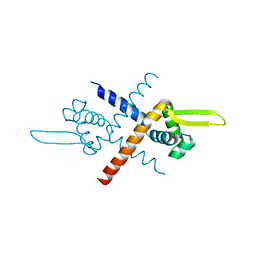 | | Structure of Bacillus subtilis HxlR, wild type | | Descriptor: | HTH-type transcriptional activator HxlR | | Authors: | Zhu, R, Chen, P.R. | | Deposit date: | 2020-04-27 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.612 Å) | | Cite: | Genetically encoded formaldehyde sensors inspired by a protein intra-helical crosslinking reaction.
Nat Commun, 12, 2021
|
|
7BZE
 
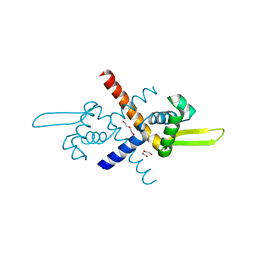 | | Structure of Bacillus subtilis HxlR, K13A mutant | | Descriptor: | DI(HYDROXYETHYL)ETHER, HTH-type transcriptional activator HxlR | | Authors: | Zhu, R, Chen, P.R. | | Deposit date: | 2020-04-27 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.658 Å) | | Cite: | Genetically encoded formaldehyde sensors inspired by a protein intra-helical crosslinking reaction.
Nat Commun, 12, 2021
|
|
7BW4
 
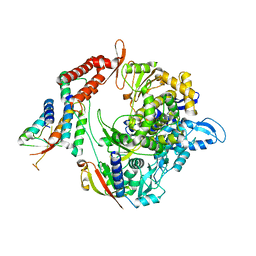 | | Structure of the RNA-dependent RNA polymerase from SARS-CoV-2 | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA-directed RNA polymerase, ... | | Authors: | Peng, Q, Peng, R, Shi, Y. | | Deposit date: | 2020-04-13 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural and Biochemical Characterization of the nsp12-nsp7-nsp8 Core Polymerase Complex from SARS-CoV-2.
Cell Rep, 31, 2020
|
|
5V2R
 
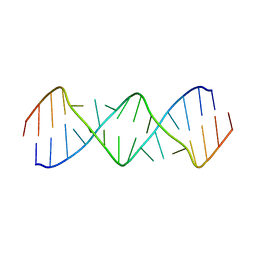 | | Structure of a GA Rich 8x8 Nucleotide RNA Internal Loop | | Descriptor: | RNA (5'-R(*CP*CP*AP*GP*AP*AP*AP*CP*GP*GP*AP*UP*GP*GP*A)-3') | | Authors: | Kauffmann, A.D, Kennedy, S.D, Turner, D.H. | | Deposit date: | 2017-03-06 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Structure of an 8 8 Nucleotide RNA Internal Loop Flanked on Each Side by Three Watson-Crick Pairs and Comparison to Three-Dimensional Predictions.
Biochemistry, 56, 2017
|
|
7MCE
 
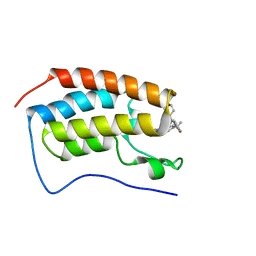 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH 2-{(7P)-7-(1,4-dimethyl-1H-1,2,3-triazol-5-yl)-8-fluoro-5-[(S)-(oxan-4-yl)(phenyl)methyl]-5H-pyrido[3,2-b]indol-3-yl}propan-2-ol | | Descriptor: | 2-{(7P)-7-(1,4-dimethyl-1H-1,2,3-triazol-5-yl)-8-fluoro-5-[(S)-(oxan-4-yl)(phenyl)methyl]-5H-pyrido[3,2-b]indol-3-yl}propan-2-ol, Bromodomain-containing protein 4 | | Authors: | Sheriff, S. | | Deposit date: | 2021-04-02 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Development of BET inhibitors as potential treatments for cancer: A new carboline chemotype.
Bioorg.Med.Chem.Lett., 51, 2021
|
|
7MK3
 
 | | Crystal structure of NPR1 | | Descriptor: | CHLORIDE ION, GLYCEROL, Regulatory protein NPR1, ... | | Authors: | Cheng, J, Wu, Q, Zhou, P. | | Deposit date: | 2021-04-21 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structural basis of NPR1 in activating plant immunity.
Nature, 605, 2022
|
|
7TAD
 
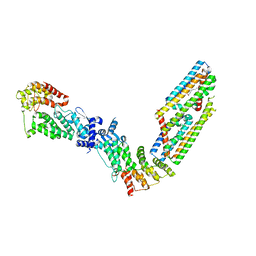 | | CryoEM structure of the (NPR1)2-(TGA3)2 complex | | Descriptor: | PALMITIC ACID, Regulatory protein NPR1, Transcription factor TGA3, ... | | Authors: | Wu, Q, Zhou, Y, Bartesaghi, A, Dong, X, Zhou, P. | | Deposit date: | 2021-12-20 | | Release date: | 2022-03-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of NPR1 in activating plant immunity.
Nature, 605, 2022
|
|
7TAC
 
 | | Cryo-EM structure of the (TGA3)2-(NPR1)2-(TGA3)2 complex | | Descriptor: | PALMITIC ACID, Regulatory protein NPR1, Transcription factor TGA3, ... | | Authors: | Wu, Q, Zhou, Y, Bartesaghi, A, Dong, X, Zhou, P. | | Deposit date: | 2021-12-20 | | Release date: | 2022-03-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of NPR1 in activating plant immunity.
Nature, 605, 2022
|
|
7TAE
 
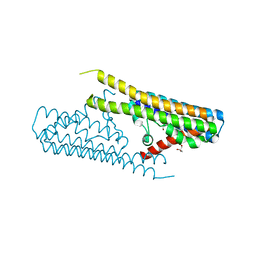 | |
6PIB
 
 | | Structure of the Klebsiella pneumoniae LpxH-AZ1 complex | | Descriptor: | 1-[5-({4-[3-(trifluoromethyl)phenyl]piperazin-1-yl}sulfonyl)-2,3-dihydro-1H-indol-1-yl]ethan-1-one, MANGANESE (II) ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Cho, J, Zhou, P. | | Deposit date: | 2019-06-26 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural basis of the UDP-diacylglucosamine pyrophosphohydrolase LpxH inhibition by sulfonyl piperazine antibiotics.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
