8H8D
 
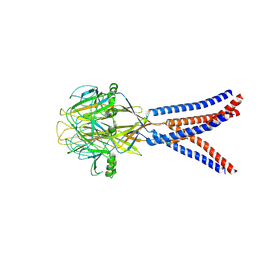 | | Structure of Xenopus tropicalis acid-sensitive outwardly rectifying channel ASOR trimer bound with tRNA (intermediate state) | | Descriptor: | Proton-activated chloride channel | | Authors: | Chi, P, Wang, X, Li, J, Li, K, Zhang, Y, Geng, J, Wu, J, Deng, D. | | Deposit date: | 2022-10-22 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.26 Å) | | Cite: | Molecular insights into the inhibition of proton-activated chloride channel by transfer RNA.
Cell Res., 34, 2024
|
|
8J57
 
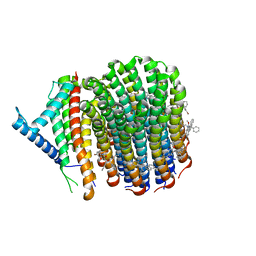 | | Cryo-EM structure of Mycobacterium tuberculosis ATP synthase Fo in complex with bedaquiline(BDQ) | | Descriptor: | ATP synthase subunit a, ATP synthase subunit c, Bedaquiline | | Authors: | Zhang, Y, Lai, Y, Liu, F, Rao, Z, Gong, H. | | Deposit date: | 2023-04-21 | | Release date: | 2024-05-01 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Inhibition of M. tuberculosis and human ATP synthase by BDQ and TBAJ-587.
Nature, 631, 2024
|
|
8J58
 
 | | Cryo-EM structure of Mycobacterium tuberculosis ATP synthase Fo in the apo-form | | Descriptor: | ATP synthase subunit a, ATP synthase subunit c | | Authors: | Zhang, Y, Lai, Y, Liu, F, Rao, Z, Gong, H. | | Deposit date: | 2023-04-21 | | Release date: | 2024-05-01 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Inhibition of M. tuberculosis and human ATP synthase by BDQ and TBAJ-587.
Nature, 631, 2024
|
|
8J0S
 
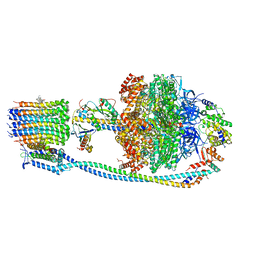 | | Cryo-EM structure of Mycobacterium tuberculosis ATP synthase in complex with bedaquiline(BDQ) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Zhang, Y, Lai, Y, Liu, F, Rao, Z, Gong, H. | | Deposit date: | 2023-04-11 | | Release date: | 2024-05-22 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Inhibition of M. tuberculosis and human ATP synthase by BDQ and TBAJ-587.
Nature, 631, 2024
|
|
5GZA
 
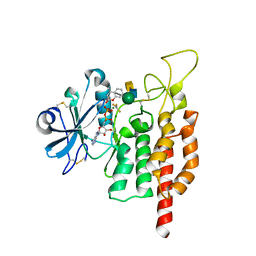 | | protein O-mannose kinase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-mannopyranose, 4-METHYL-2H-CHROMEN-2-ONE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Xiao, J. | | Deposit date: | 2016-09-27 | | Release date: | 2016-12-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of protein O-mannose kinase reveals a unique active site architecture
Elife, 5, 2016
|
|
8JR0
 
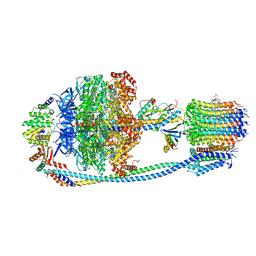 | | Cryo-EM structure of Mycobacterium tuberculosis ATP synthase in complex with TBAJ-587 | | Descriptor: | (1~{S},2~{S})-1-(6-bromanyl-2-methoxy-quinolin-3-yl)-2-(2,6-dimethoxypyridin-4-yl)-4-(dimethylamino)-1-(2-fluoranyl-3-methoxy-phenyl)butan-2-ol, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhang, Y, Lai, Y, Liu, F, Rao, Z, Gong, H. | | Deposit date: | 2023-06-15 | | Release date: | 2024-05-01 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Inhibition of M. tuberculosis and human ATP synthase by BDQ and TBAJ-587.
Nature, 631, 2024
|
|
8KHF
 
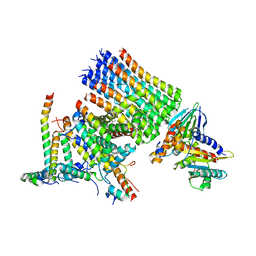 | |
8JR1
 
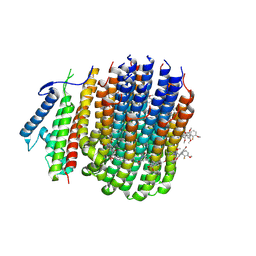 | | Cryo-EM structure of Mycobacterium tuberculosis ATP synthase Fo in complex with TBAJ-587 | | Descriptor: | (1~{S},2~{S})-1-(6-bromanyl-2-methoxy-quinolin-3-yl)-2-(2,6-dimethoxypyridin-4-yl)-4-(dimethylamino)-1-(2-fluoranyl-3-methoxy-phenyl)butan-2-ol, ATP synthase subunit a, ATP synthase subunit c | | Authors: | Zhang, Y, Lai, Y, Liu, F, Rao, Z, Gong, H. | | Deposit date: | 2023-06-15 | | Release date: | 2024-05-01 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Inhibition of M. tuberculosis and human ATP synthase by BDQ and TBAJ-587.
Nature, 631, 2024
|
|
8KI3
 
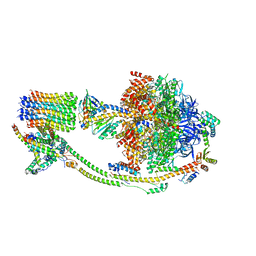 | | Structure of the human ATP synthase bound to bedaquiline (composite) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase F(0) complex subunit B1, ... | | Authors: | Lai, Y, Zhang, Y, Gong, H. | | Deposit date: | 2023-08-22 | | Release date: | 2024-05-01 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Inhibition of M. tuberculosis and human ATP synthase by BDQ and TBAJ-587.
Nature, 631, 2024
|
|
8J0T
 
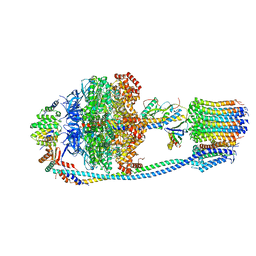 | | Cryo-EM structure of Mycobacterium tuberculosis ATP synthase in the apo-form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Zhang, Y, Lai, Y, Liu, F, Rao, Z, Gong, H. | | Deposit date: | 2023-04-11 | | Release date: | 2024-05-22 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Inhibition of M. tuberculosis and human ATP synthase by BDQ and TBAJ-587.
Nature, 631, 2024
|
|
6KYL
 
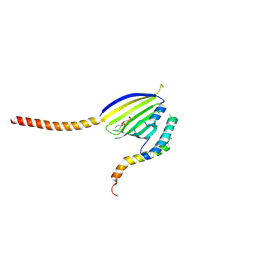 | | Crystal Structure of Phosphatidic acid Transporter Ups1/Mdm35 in Complex with (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate, Mitochondrial distribution and morphology protein 35, Protein UPS1, ... | | Authors: | Lu, J, Chan, K.C, Zhai, Y, Fan, J, Sun, F. | | Deposit date: | 2019-09-19 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Molecular mechanism of mitochondrial phosphatidate transfer by Ups1.
Commun Biol, 3, 2020
|
|
6LUM
 
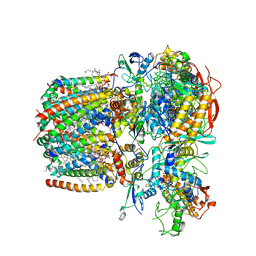 | | Structure of Mycobacterium smegmatis succinate dehydrogenase 2 | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOINOSITOL, 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, ... | | Authors: | Gao, Y, Gong, H, Zhou, X, Xiao, Y, Wang, W, Ji, W, Wang, Q, Rao, Z. | | Deposit date: | 2020-01-29 | | Release date: | 2020-05-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Cryo-EM structure of trimeric Mycobacterium smegmatis succinate dehydrogenase with a membrane-anchor SdhF.
Nat Commun, 11, 2020
|
|
8DQK
 
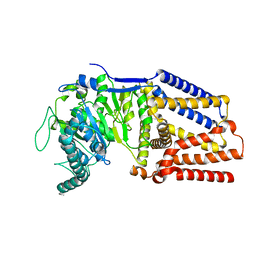 | | Intermediate resolution structure of barley (1,3;1,4)-beta-glucan synthase CslF6. | | Descriptor: | Cellulose synthase-like CslF6 | | Authors: | Ho, R, Purushotham, P, Zimmer, J. | | Deposit date: | 2022-07-19 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Mechanism of mixed-linkage glucan biosynthesis by barley cellulose synthase-like CslF6 (1,3;1,4)-beta-glucan synthase.
Sci Adv, 8, 2022
|
|
8D89
 
 | | Crystal structure of a novel GH5 enzyme retrieved from capybara gut metagenome | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Martins, M.P, Morais, M.A.B, Murakami, M.T. | | Deposit date: | 2022-06-08 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Glycoside hydrolase subfamily GH5_57 features a highly redesigned catalytic interface to process complex hetero-beta-mannans.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
4N0O
 
 | | Complex structure of Arterivirus nonstructural protein 10 (helicase) with DNA | | Descriptor: | CALCIUM ION, DNA, Replicase polyprotein 1ab, ... | | Authors: | Deng, Z, Chen, Z. | | Deposit date: | 2013-10-02 | | Release date: | 2014-01-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for the regulatory function of a complex zinc-binding domain in a replicative arterivirus helicase resembling a nonsense-mediated mRNA decay helicase.
Nucleic Acids Res., 42, 2014
|
|
4N0N
 
 | | Crystal structure of Arterivirus nonstructural protein 10 (helicase) | | Descriptor: | MAGNESIUM ION, Replicase polyprotein 1ab, SULFATE ION, ... | | Authors: | Deng, Z, Chen, Z. | | Deposit date: | 2013-10-02 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the regulatory function of a complex zinc-binding domain in a replicative arterivirus helicase resembling a nonsense-mediated mRNA decay helicase.
Nucleic Acids Res., 42, 2014
|
|
4FWE
 
 | | Native structure of LSD2 /AOF1/KDM1b in spacegroup of C2221 at 2.13A | | Descriptor: | CITRATE ANION, FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1B, ... | | Authors: | Zhang, Q, Chen, Z. | | Deposit date: | 2012-07-01 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structure-function analysis reveals a novel mechanism for regulation of histone demethylase LSD2/AOF1/KDM1b
Cell Res., 23, 2013
|
|
4FWF
 
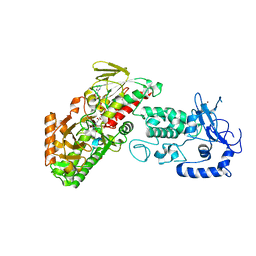 | | Complex structure of LSD2/AOF1/KDM1b with H3K4 mimic | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Histone H3.1, Lysine-specific histone demethylase 1B, ... | | Authors: | Zhang, Q, Chen, Z. | | Deposit date: | 2012-07-01 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-function analysis reveals a novel mechanism for regulation of histone demethylase LSD2/AOF1/KDM1b
Cell Res., 23, 2013
|
|
7MLJ
 
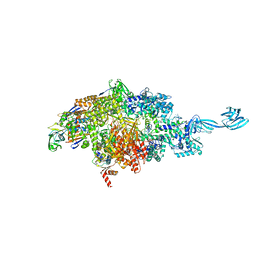 | |
7MLB
 
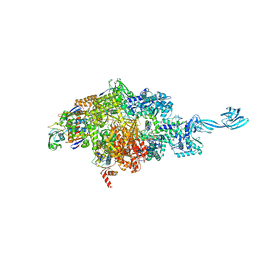 | |
7MLI
 
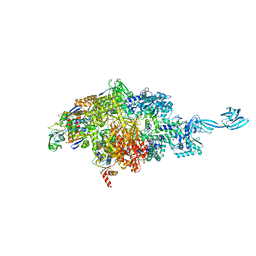 | |
4FWJ
 
 | | Native structure of LSD2/AOF1/KDM1b in spacegroup of I222 at 2.9A | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1B, PHOSPHATE ION, ... | | Authors: | Zhang, Q, Chen, Z. | | Deposit date: | 2012-07-01 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-function analysis reveals a novel mechanism for regulation of histone demethylase LSD2/AOF1/KDM1b
Cell Res., 23, 2013
|
|
7EFX
 
 | | Crystal Structure of human PIN1 complexed with covalent inhibitor | | Descriptor: | 4-((5-bromofuran-2-yl)methyl)-8-(2-chloroacetyl)-1-thia-4,8-diazaspiro[4.5]decan-3-one, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Liu, L, Li, J, Zhu, R, Pei, Y. | | Deposit date: | 2021-03-23 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Computational and Structure-Based Development of High Potent Cell-Active Covalent Inhibitor Targeting the Peptidyl-Prolyl Isomerase NIMA-Interacting-1 (Pin1).
J.Med.Chem., 65, 2022
|
|
7EKV
 
 | | Crystal Structure of human Pin1 complexed with a covalent inhibitor | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 8-(2-chloroacetyl)-4-((5-phenylfuran-2-yl)methyl)-1-thia-4,8-diazaspiro[4.5]decan-3-one, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Liu, L, Li, J. | | Deposit date: | 2021-04-06 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Computational and Structure-Based Development of High Potent Cell-Active Covalent Inhibitor Targeting the Peptidyl-Prolyl Isomerase NIMA-Interacting-1 (Pin1).
J.Med.Chem., 65, 2022
|
|
7EFJ
 
 | | Crystal Structure Analysis of human PIN1 | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 8-(2-chloroacetyl)-4-(furan-2-ylmethyl)-1-thia-4,8-diazaspiro[4.5]decan-3-one, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Liu, L, Li, J. | | Deposit date: | 2021-03-21 | | Release date: | 2022-02-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Computational and Structure-Based Development of High Potent Cell-Active Covalent Inhibitor Targeting the Peptidyl-Prolyl Isomerase NIMA-Interacting-1 (Pin1).
J.Med.Chem., 65, 2022
|
|
