8B21
 
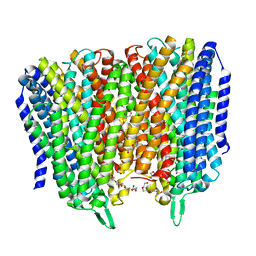 | | Time-resolved structure of K+-dependent Na+-PPase from Thermotoga maritima 0-60-seconds post reaction initiation with Na+ | | Descriptor: | DI(HYDROXYETHYL)ETHER, DODECYL-BETA-D-MALTOSIDE, K(+)-stimulated pyrophosphate-energized sodium pump, ... | | Authors: | Strauss, J, Vidilaseris, K, Goldman, A. | | Deposit date: | 2022-09-12 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Functional and structural asymmetry suggest a unifying principle for catalysis in membrane-bound pyrophosphatases.
Embo Rep., 25, 2024
|
|
8B24
 
 | | Time-resolved structure of K+-dependent Na+-PPase from Thermotoga maritima 3600-seconds post reaction initiation with Na+ | | Descriptor: | DIPHOSPHATE, K(+)-stimulated pyrophosphate-energized sodium pump, MAGNESIUM ION, ... | | Authors: | Strauss, J, Vidilaseris, K, Goldman, A. | | Deposit date: | 2022-09-12 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (4.53 Å) | | Cite: | Functional and structural asymmetry suggest a unifying principle for catalysis in membrane-bound pyrophosphatases.
Embo Rep., 25, 2024
|
|
8CD8
 
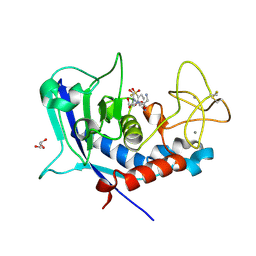 | | Ulilysin - C269A with AEBSF complex | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, CALCIUM ION, GLY-SER-SER, ... | | Authors: | Rodriguez-Banqueri, A, Eckhard, U, Gomis-Ruth, F.X. | | Deposit date: | 2023-01-30 | | Release date: | 2023-03-22 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into latency of the metallopeptidase ulilysin (lysargiNase) and its unexpected inhibition by a sulfonyl-fluoride inhibitor of serine peptidases.
Dalton Trans, 52, 2023
|
|
8C3P
 
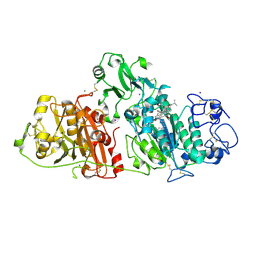 | |
8C3O
 
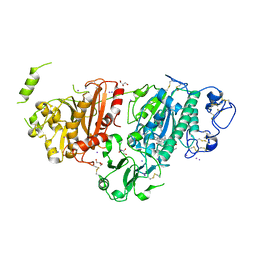 | | Crystal structure of autotaxin gamma and compound MEY-003 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5,7-bis(oxidanyl)-2-(1-pentylindol-3-yl)chromen-4-one, 7alpha-hydroxycholesterol, ... | | Authors: | Eymery, M.C, McCarthy, A.A. | | Deposit date: | 2022-12-27 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Discovery of potent chromone-based autotaxin inhibitors inspired by cannabinoids.
Eur.J.Med.Chem., 263, 2023
|
|
8C4W
 
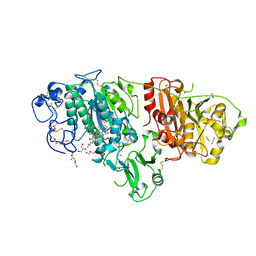 | | Crystal structure of rat autotaxin and compound MEY-002 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5,7-bis(oxidanyl)-2-[1-(phenylmethyl)indol-3-yl]chromen-4-one, 7alpha-hydroxycholesterol, ... | | Authors: | Eymery, M.C, McCarthy, A.A. | | Deposit date: | 2023-01-05 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of potent chromone-based autotaxin inhibitors inspired by cannabinoids.
Eur.J.Med.Chem., 263, 2023
|
|
8C7R
 
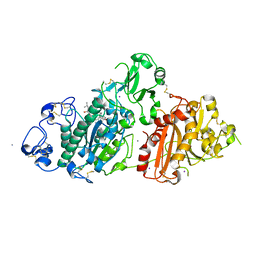 | | Crystal structure of rat autotaxin and compound MEY-003 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5,7-bis(oxidanyl)-2-(1-pentylindol-3-yl)chromen-4-one, 7alpha-hydroxycholesterol, ... | | Authors: | Eymery, M.C, McCarthy, A.A. | | Deposit date: | 2023-01-17 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Discovery of potent chromone-based autotaxin inhibitors inspired by cannabinoids.
Eur.J.Med.Chem., 263, 2023
|
|
8C3T
 
 | |
8CGA
 
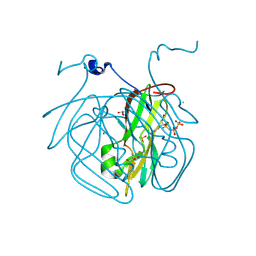 | | Structure of Mycobacterium tuberculosis dUTPase delta 133A-137S mutant | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Toth, Z.S, Benedek, A, Leveles, I, Vertessy, B.G. | | Deposit date: | 2023-02-03 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of Mycobacterium tuberculosis dUTPase delta 133A-137S mutant
To Be Published
|
|
8DAF
 
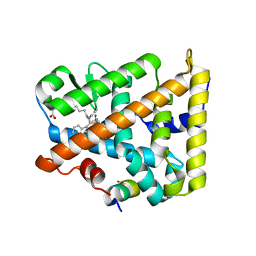 | | Human SF-1 LBD bound to synthetic agonist 6N-10CA and bacterial phospholipid | | Descriptor: | 10-[(3aR,6S,6aR)-3-phenyl-3a-(1-phenylethenyl)-6-(sulfamoylamino)-1,3a,4,5,6,6a-hexahydropentalen-2-yl]decanoic acid (non-preferred name), DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, Nuclear receptor coactivator 2, ... | | Authors: | D'Agostino, E.H, Cato, M.L, Ortlund, E.A. | | Deposit date: | 2022-06-13 | | Release date: | 2023-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Comparison of activity, structure, and dynamics of SF-1 and LRH-1 complexed with small molecule modulators.
J.Biol.Chem., 299, 2023
|
|
8D86
 
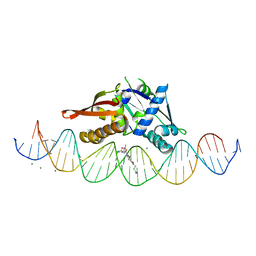 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence co-crystallized with a guest small molecule, netropsin. | | Descriptor: | DNA (5'-D(A*CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*G)-3'), DNA (5'-D(A*GP*GP*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*G)-3'), MAGNESIUM ION, ... | | Authors: | Orun, A.R, Snow, C.D. | | Deposit date: | 2022-06-07 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Modular Protein-DNA Cocrystals as Precise, Programmable Assembly Scaffolds.
Acs Nano, 17, 2023
|
|
8D8M
 
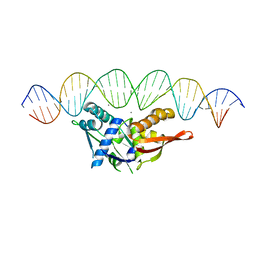 | |
8BRX
 
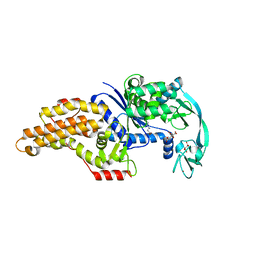 | | Escherichia coli methionyl-tRNA synthetase mutant L13C,I297C complexed with beta-3-methionine | | Descriptor: | (3R)-3-amino-5-(methylsulfanyl)pentanoic acid, CITRIC ACID, Methionine--tRNA ligase, ... | | Authors: | Schmitt, E, Mechulam, Y, Nigro, G, Opuu, V, Lazennec-Schurdevin, C, Simonson, T. | | Deposit date: | 2022-11-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Redesigning methionyl-tRNA synthetase for beta-methionine activity with adaptive landscape flattening and experiments.
Protein Sci., 32, 2023
|
|
8B8H
 
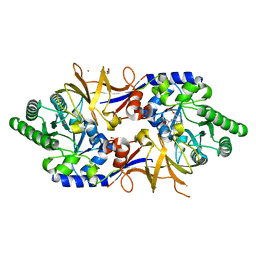 | | Structure of DCS-resistant variant D322N of alanine racemase from M. tuberculosis in complex with DCS | | Descriptor: | (~{E})-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylidene-[(4~{R})-3-oxidanylidene-1,2-oxazolidin-4-yl]azanium, 1,2-ETHANEDIOL, Alanine racemase, ... | | Authors: | de Chiara, C, Prosser, G, Ogrodowicz, R.W, de Carvalho, L.P.S. | | Deposit date: | 2022-10-04 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of the d-Cycloserine-Resistant Variant D322N of Alanine Racemase from Mycobacterium tuberculosis .
Acs Bio Med Chem Au, 3, 2023
|
|
8CEM
 
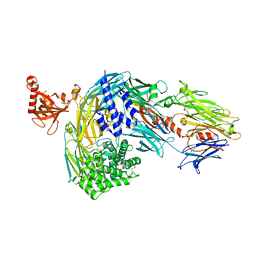 | | Structure of bovine native C3, re-refinement | | Descriptor: | Complement C3 alpha chain, Complement C3 beta chain, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Andersen, G.R, Fredslund, F. | | Deposit date: | 2023-02-02 | | Release date: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of bovine complement component 3 reveals the basis for thioester function.
J Mol Biol, 361, 2006
|
|
2SNS
 
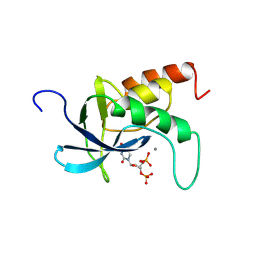 | | STAPHYLOCOCCAL NUCLEASE. PROPOSED MECHANISM OF ACTION BASED ON STRUCTURE OF ENZYME-THYMIDINE 3(PRIME),5(PRIME)-BIPHOSPHATE-CALCIUM ION COMPLEX AT 1.5-ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, THERMONUCLEASE PRECURSOR, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Legg, M.J, Cotton, F.A, Hazen Jr, E.E. | | Deposit date: | 1982-05-14 | | Release date: | 1982-07-29 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: |
Thesis, Texas Agricultural and Mechanical University, 1977
|
|
1VK1
 
 | | Conserved hypothetical protein from Pyrococcus furiosus Pfu-392566-001 | | Descriptor: | Conserved hypothetical protein, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Shah, A, Liu, Z.J, Tempel, W, Chen, L, Lee, D, Yang, H, Chang, J, Zhao, M, Ng, J, Rose, J, Brereton, P.S, Izumi, M, Jenney Jr, F.E, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-04-13 | | Release date: | 2004-08-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | (NZ)CH...O contacts assist crystallization of a ParB-like nuclease.
Bmc Struct.Biol., 7, 2007
|
|
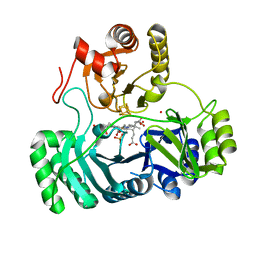
1AOP
 
 | |
8QZO
 
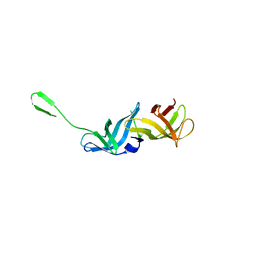 | |
8QU4
 
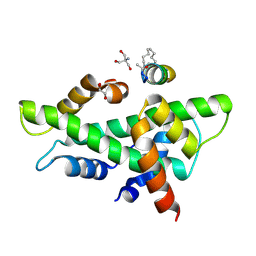 | | NF-YB/C Heterodimer in Complex with a 13-mer NF-YA-derived Peptide Stabilized with C8-Hydrocarbon Linker in an alternative binding pose | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Nuclear transcription factor Y subunit alpha, ... | | Authors: | Arbore, F, Durukan, C, Klintrot, C.I.R, Grossmann, T.N, Hennig, S. | | Deposit date: | 2023-10-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Binding Dynamics of a Stapled Peptide Targeting the Transcription Factor NF-Y.
Chembiochem, 25, 2024
|
|
8QU2
 
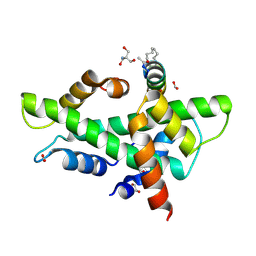 | | NF-YB/C Heterodimer in Complex with a 16-mer NF-YA-derived Peptide Stabilized with C8-Hydrocarbon Linker | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, ... | | Authors: | Durukan, C, Arbore, F, Klintrot, C.I.R, Grossmann, T.N, Hennig, S. | | Deposit date: | 2023-10-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Binding Dynamics of a Stapled Peptide Targeting the Transcription Factor NF-Y.
Chembiochem, 25, 2024
|
|
8QU3
 
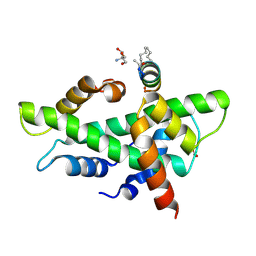 | | NF-YB/C Heterodimer in Complex with a 13-mer NF-YA-derived Peptide Stabilized with C8-Hydrocarbon Linker | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, Nuclear transcription factor Y subunit alpha, ... | | Authors: | Arbore, F, Durukan, C, Klintrot, C.I.R, Grossmann, T.N, Hennig, S. | | Deposit date: | 2023-10-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Binding Dynamics of a Stapled Peptide Targeting the Transcription Factor NF-Y.
Chembiochem, 25, 2024
|
|
8RZE
 
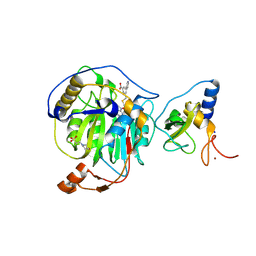 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 10 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]-5-pyridin-3-yl-benzoic acid, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-02-12 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 nsp16 by substrate-based dual site inhibitors.
Chemmedchem, 2024
|
|
8RZC
 
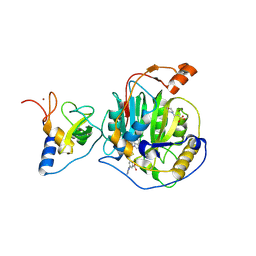 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 11 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]-5-imidazol-1-yl-benzoic acid, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-02-12 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 nsp16 by substrate-based dual site inhibitors.
Chemmedchem, 2024
|
|
8RZD
 
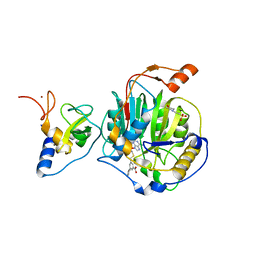 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 9 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]-5-(3-hydroxyphenyl)benzoic acid, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-02-12 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 nsp16 by substrate-based dual site inhibitors.
Chemmedchem, 2024
|
|
