3HUP
 
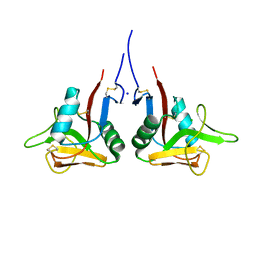 | | High-resolution structure of the extracellular domain of human CD69 | | Descriptor: | CHLORIDE ION, Early activation antigen CD69, SODIUM ION | | Authors: | Kolenko, P, Dohnalek, J, Skalova, T, Hasek, J, Duskova, J, Vanek, O, Bezouska, K. | | Deposit date: | 2009-06-15 | | Release date: | 2009-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.371 Å) | | Cite: | The high-resolution structure of the extracellular domain of human CD69 using a novel polymer
Acta Crystallogr.,Sect.F, 65, 2009
|
|
7TLU
 
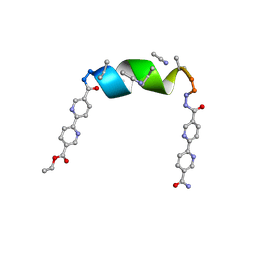 | |
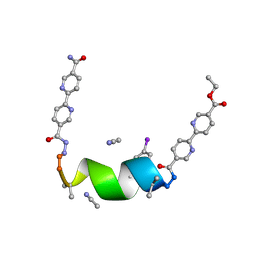
7TMH
 
 | |
7TML
 
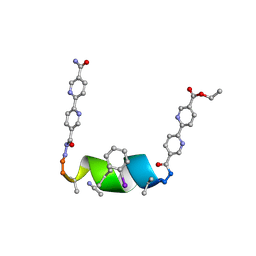 | |
7TMK
 
 | |
7TLS
 
 | |
7TM2
 
 | |
7TMJ
 
 | |
6GJV
 
 | |
7TM1
 
 | |
7TMI
 
 | |
6E7H
 
 | |
6SJT
 
 | |
6SKW
 
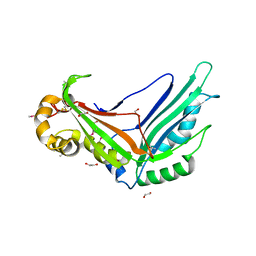 | | Crystal structure of the Legionella pneumophila type II secretion system substrate NttE | | Descriptor: | 1,2-ETHANEDIOL, NttE | | Authors: | Portlock, T.J, Rehman, S, Garnett, J.A. | | Deposit date: | 2019-08-16 | | Release date: | 2020-05-20 | | Last modified: | 2020-07-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure, Dynamics and Cellular Insight Into Novel Substrates of theLegionella pneumophilaType II Secretion System.
Front Mol Biosci, 7, 2020
|
|
3FPD
 
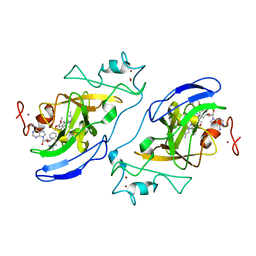 | | G9a-like protein lysine methyltransferase inhibition by BIX-01294 | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-9 specific 5, N-(1-benzylpiperidin-4-yl)-6,7-dimethoxy-2-(4-methyl-1,4-diazepan-1-yl)quinazolin-4-amine, ... | | Authors: | Chang, Y, Zhang, X, Horton, J.R, Cheng, X. | | Deposit date: | 2009-01-05 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for G9a-like protein lysine methyltransferase inhibition by BIX-01294.
Nat.Struct.Mol.Biol., 16, 2009
|
|
2O6L
 
 | |
7UIB
 
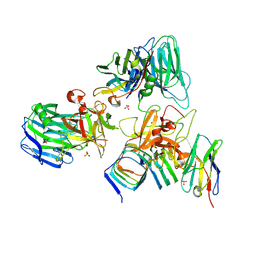 | | Crystal structure of BoNT/E receptor binding domain in complex with SV2, VHH, and sialic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetyl-beta-neuraminic acid, ... | | Authors: | Liu, Z, Jin, R, Chen, P. | | Deposit date: | 2022-03-29 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural basis for botulinum neurotoxin E recognition of synaptic vesicle protein 2.
Nat Commun, 14, 2023
|
|
3CG8
 
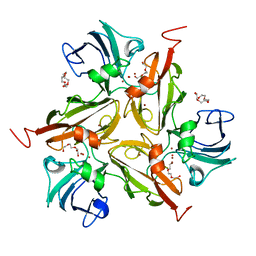 | | Laccase from Streptomyces coelicolor | | Descriptor: | COPPER (II) ION, OXYGEN ATOM, TETRAETHYLENE GLYCOL, ... | | Authors: | Skalova, T, Dohnalek, J, Ostergaard, L.H, Ostergaard, P.R, Kolenko, P, Duskova, J, Hasek, J. | | Deposit date: | 2008-03-05 | | Release date: | 2009-01-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.679 Å) | | Cite: | The Structure of the Small Laccase from Streptomyces coelicolor Reveals a Link between Laccases and Nitrite Reductases.
J.Mol.Biol., 385, 2009
|
|
7UIA
 
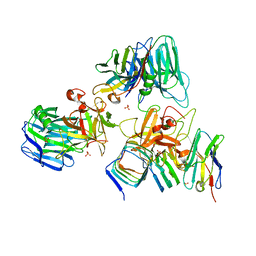 | | Crystal structure of BoNT/E receptor binding domain in complex with SV2 and VHH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Liu, Z, Jin, R, Chen, P. | | Deposit date: | 2022-03-28 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural basis for botulinum neurotoxin E recognition of synaptic vesicle protein 2.
Nat Commun, 14, 2023
|
|
7UIE
 
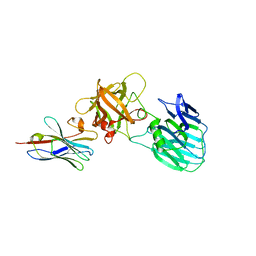 | | Crystal structure of HcE-JLE-G6 | | Descriptor: | Botulinum neurotoxin E heavy chain, JLE-G6 | | Authors: | Jin, R, Lam, K. | | Deposit date: | 2022-03-29 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Structural basis for botulinum neurotoxin E recognition of synaptic vesicle protein 2.
Nat Commun, 14, 2023
|
|
6E7G
 
 | |
3RNX
 
 | |
3RS1
 
 | | Mouse C-type lectin-related protein Clrg | | Descriptor: | C-type lectin domain family 2 member I, CHLORIDE ION | | Authors: | Skalova, T, Dohnalek, J, Duskova, J, Stepankova, A, Koval, T, Hasek, J, Kotynkova, K, Vanek, O, Bezouska, K. | | Deposit date: | 2011-05-02 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mouse Clr-g, a Ligand for NK Cell Activation Receptor NKR-P1F: Crystal Structure and Biophysical Properties.
J.Immunol., 189, 2012
|
|
5MY5
 
 | |
2O8M
 
 | | Crystal structure of the S139A mutant of Hepatitis C Virus NS3/4A protease | | Descriptor: | Protease, SODIUM ION, ZINC ION | | Authors: | Fischmann, T.O, Prongay, A.J, Madison, V.M, Yao, N. | | Deposit date: | 2006-12-12 | | Release date: | 2007-10-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization
J.Med.Chem., 50, 2007
|
|
