5Y8J
 
 | | Mycobacterium tuberculosis 3-Hydroxyisobutyrate dehydrogenase (MtHIBADH) + (R)-3-hydroxyisobutyrate (R-HIBA) | | Descriptor: | (2R)-3-HYDROXY-2-METHYLPROPANOIC ACID, (2~{S})-2-methylpentanedioic acid, ACRYLIC ACID, ... | | Authors: | Srikalaivani, R, Singh, A, Surolia, A, Vijayan, M. | | Deposit date: | 2017-08-21 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure, interactions and action ofMycobacterium tuberculosis3-hydroxyisobutyric acid dehydrogenase.
Biochem. J., 475, 2018
|
|
5Y8L
 
 | | Mycobacterium tuberculosis 3-Hydroxyisobutyrate dehydrogenase (MtHIBADH) + NAD +(S)-3-hydroxyisobutyrate (S-HIBA) | | Descriptor: | (2S)-2-methyl-3-oxidanyl-propanoic acid, (2~{S})-2-methylpentanedioic acid, GLYCEROL, ... | | Authors: | Srikalaivani, R, Singh, A, Surolia, A, Vijayan, M. | | Deposit date: | 2017-08-21 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure, interactions and action ofMycobacterium tuberculosis3-hydroxyisobutyric acid dehydrogenase.
Biochem. J., 475, 2018
|
|
5Y8N
 
 | | Mycobacterium tuberculosis 3-Hydroxyisobutyrate dehydrogenase (MtHIBADH) + NAD + L-serine | | Descriptor: | (2~{S})-2-methylpentanedioic acid, ACRYLIC ACID, GLYCEROL, ... | | Authors: | Srikalaivani, R, Singh, A, Surolia, A, Vijayan, M. | | Deposit date: | 2017-08-21 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structure, interactions and action ofMycobacterium tuberculosis3-hydroxyisobutyric acid dehydrogenase.
Biochem. J., 475, 2018
|
|
5Y8H
 
 | | Mycobacterium tuberculosis 3-Hydroxyisobutyrate dehydrogenase (MtHIBADH) + NAD+ | | Descriptor: | (2~{S})-2-methylpentanedioic acid, ACRYLIC ACID, GLYCEROL, ... | | Authors: | Srikalaivani, R, Singh, A, Surolia, A, Vijayan, M. | | Deposit date: | 2017-08-21 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure, interactions and action ofMycobacterium tuberculosis3-hydroxyisobutyric acid dehydrogenase.
Biochem. J., 475, 2018
|
|
8UCE
 
 | | Thermophilic RNA Ligase from Palaeococcus pacificus + AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP dependent DNA ligase, GLYCEROL, ... | | Authors: | Rousseau, M.D, Hicks, J.L, Oulavallickal, T, Williamson, A, Arcus, V.L, Patrick, M.W. | | Deposit date: | 2023-09-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Characterisation and engineering of a thermophilic RNA ligase from Palaeococcus pacificus.
Nucleic Acids Res., 52, 2024
|
|
8UCI
 
 | | Thermophilic RNA Ligase from Palaeococcus pacificus K238G + AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP dependent DNA ligase, GLYCEROL, ... | | Authors: | Rousseau, M.D, Hicks, J.L, Oulavallickal, T, Williamson, A, Arcus, V.L, Patrick, M.W. | | Deposit date: | 2023-09-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Characterisation and engineering of a thermophilic RNA ligase from Palaeococcus pacificus.
Nucleic Acids Res., 52, 2024
|
|
8UCG
 
 | | Thermophilic RNA Ligase from Palaeococcus pacificus K92A | | Descriptor: | ATP dependent DNA ligase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Rousseau, M.D, Hicks, J.L, Oulavallickal, T, Williamson, A, Arcus, V.L, Patrick, M.W. | | Deposit date: | 2023-09-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Characterisation and engineering of a thermophilic RNA ligase from Palaeococcus pacificus.
Nucleic Acids Res., 52, 2024
|
|
8UCH
 
 | | Thermophilic RNA Ligase from Palaeococcus pacificus K92A + ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP dependent DNA ligase, GLYCEROL, ... | | Authors: | Rousseau, M.D, Hicks, J.L, Oulavallickal, T, Williamson, A, Arcus, V.L, Patrick, M.W. | | Deposit date: | 2023-09-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Characterisation and engineering of a thermophilic RNA ligase from Palaeococcus pacificus.
Nucleic Acids Res., 52, 2024
|
|
8UCF
 
 | | Thermophilic RNA Ligase from Palaeococcus pacificus K238G | | Descriptor: | ATP dependent DNA ligase, GLYCEROL, MAGNESIUM ION | | Authors: | Rousseau, M.D, Hicks, J.L, Oulavallickal, T, Williamson, A, Arcus, V.L, Patrick, M.W. | | Deposit date: | 2023-09-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterisation and engineering of a thermophilic RNA ligase from Palaeococcus pacificus.
Nucleic Acids Res., 52, 2024
|
|
8TON
 
 | |
5Y8O
 
 | | Mycobacterium tuberculosis 3-Hydroxyisobutyrate dehydrogenase (MtHIBADH) + NAD + 3-Hydroxy propionate (3-HP) | | Descriptor: | (2~{S})-2-methylpentanedioic acid, 3-HYDROXY-PROPANOIC ACID, ACRYLIC ACID, ... | | Authors: | Srikalaivani, R, Singh, A, Surolia, A, Vijayan, M. | | Deposit date: | 2017-08-21 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure, interactions and action ofMycobacterium tuberculosis3-hydroxyisobutyric acid dehydrogenase.
Biochem. J., 475, 2018
|
|
5Y8I
 
 | | Mycobacterium tuberculosis 3-Hydroxyisobutyrate dehydrogenase (MtHIBADH) + (S)-3-hydroxyisobutyrate (S-HIBA) | | Descriptor: | (2S)-2-methyl-3-oxidanyl-propanoic acid, (2~{S})-2-methylpentanedioic acid, GLYCEROL, ... | | Authors: | Srikalaivani, R, Singh, A, Surolia, A, Vijayan, M. | | Deposit date: | 2017-08-21 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure, interactions and action ofMycobacterium tuberculosis3-hydroxyisobutyric acid dehydrogenase.
Biochem. J., 475, 2018
|
|
5Y8P
 
 | | Mycobacterium tuberculosis 3-Hydroxyisobutyrate dehydrogenase (MtHIBADH) + 3-Hydroxy propionate (3-HP) | | Descriptor: | (2~{S})-2-methylpentanedioic acid, 3-HYDROXY-PROPANOIC ACID, ACRYLIC ACID, ... | | Authors: | Srikalaivani, R, Singh, A, Surolia, A, Vijayan, M. | | Deposit date: | 2017-08-21 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure, interactions and action ofMycobacterium tuberculosis3-hydroxyisobutyric acid dehydrogenase.
Biochem. J., 475, 2018
|
|
2O4K
 
 | | Crystal Structure of HIV-1 Protease (Q7K) in Complex with Atazanavir | | Descriptor: | (3S,8S,9S,12S)-3,12-BIS(1,1-DIMETHYLETHYL)-8-HYDROXY-4,11-DIOXO-9-(PHENYLMETHYL)-6-[[4-(2-PYRIDINYL)PHENYL]METHYL]-2,5, 6,10,13-PENTAAZATETRADECANEDIOIC ACID DIMETHYL ESTER, CHLORIDE ION, ... | | Authors: | Armstrong, A.A, Muzammil, S, Jakalian, A, Bonneau, P.R, Schmelmer, V, Freire, E, Amzel, L.M. | | Deposit date: | 2006-12-04 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Unique thermodynamic response of tipranavir to human immunodeficiency virus type 1 protease drug resistance mutations.
J.Virol., 81, 2007
|
|
2O4L
 
 | | Crystal Structure of HIV-1 Protease (Q7K, I50V) in Complex with Tipranavir | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(3-{(1R)-1-[(6R)-4-HYDROXY-2-OXO-6-PHENETHYL-6-PROPYL-5,6-DIHYDRO-2H-PYRAN-3-YL]PROPYL}PHENYL)-5-(TRIFLUOROMETHYL)-2-PYRIDINESULFONAMIDE, ... | | Authors: | Armstrong, A.A, Muzammil, S, Jakalian, A, Bonneau, P.R, Schmelmer, V, Freire, E, Amzel, L.M. | | Deposit date: | 2006-12-04 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Unique thermodynamic response of tipranavir to human immunodeficiency virus type 1 protease drug resistance mutations.
J.Virol., 81, 2007
|
|
2O4S
 
 | | Crystal Structure of HIV-1 Protease (Q7K) in Complex with Lopinavir | | Descriptor: | CHLORIDE ION, GLYCEROL, N-{1-BENZYL-4-[2-(2,6-DIMETHYL-PHENOXY)-ACETYLAMINO]-3-HYDROXY-5-PHENYL-PENTYL}-3-METHYL-2-(2-OXO-TETRAHYDRO-PYRIMIDIN-1-YL)-BUTYRAMIDE, ... | | Authors: | Armstrong, A.A, Muzammil, S, Jakalian, A, Bonneau, P.R, Schmelmer, V, Freire, E, Amzel, L.M. | | Deposit date: | 2006-12-04 | | Release date: | 2006-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Unique thermodynamic response of tipranavir to human immunodeficiency virus type 1 protease drug resistance mutations.
J.Virol., 81, 2007
|
|
5Y97
 
 | | Crystal structure of snake gourd seed lectin in complex with lactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Seed lectin, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Chandran, T, Vijayan, M, Sivaji, N, Surolia, A. | | Deposit date: | 2017-08-23 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Ligand binding and retention in snake gourd seed lectin (SGSL). A crystallographic, thermodynamic and molecular dynamics study
Glycobiology, 28, 2018
|
|
4M65
 
 | | In situ thermolysin crystallized on a MiTeGen micromesh with asparagine ligand | | Descriptor: | 1,2-ETHANEDIOL, ASPARAGINE, CALCIUM ION, ... | | Authors: | Yin, X, Scalia, A, Leroy, L, Cuttitta, C.M, Polizzo, G.M, Ericson, D.L, Roessler, C.G, Campos, O, Agarwal, R, Allaire, M, Orville, A.M, Jackimowicz, R, Ma, M.Y, Sweet, R.M, Soares, A.S. | | Deposit date: | 2013-08-08 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Hitting the target: fragment screening with acoustic in situ
co-crystallization of proteins plus fragment libraries on
pin-mounted data-collection micromeshes
Acta Crystallogr.,Sect.D, D70
|
|
4MQ3
 
 | |
7V4Z
 
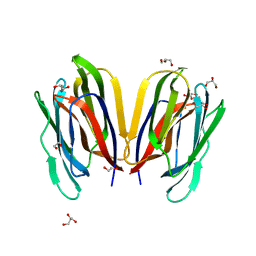 | | Structure of Horcolin native form | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Bobbili, K.B, Sivaji, N, Jayaprakash, N.G, Narayanan, V, Sekhar, A, Suguna, K, Surolia, A. | | Deposit date: | 2021-08-16 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structure and Carbohydrate Recognition by the Nonmitogenic Lectin Horcolin.
Biochemistry, 61, 2022
|
|
7V4S
 
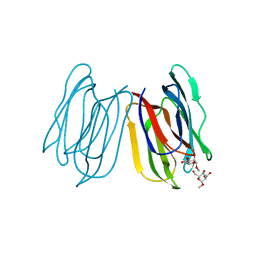 | | Horcolin complex with methyl-alpha-mannose | | Descriptor: | Horcolin, methyl alpha-D-mannopyranoside | | Authors: | Bobbili, K.B, Sivaji, N, Jayaprakash, N.G, Narayanan, V, Sekhar, A, Suguna, K, Surolia, A. | | Deposit date: | 2021-08-14 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure and Carbohydrate Recognition by the Nonmitogenic Lectin Horcolin.
Biochemistry, 61, 2022
|
|
4P2E
 
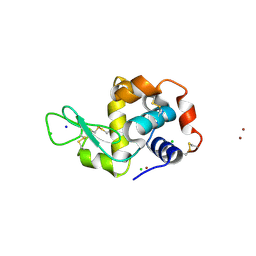 | | Acoustic transfer of protein crystals from agar pedestals to micromeshes for high throughput screening of heavy atom derivatives | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Lysozyme C, ... | | Authors: | Cuttitta, C.M, Roessler, C.G, Ericson, D.L, Scalia, A, Teplitsky, E, Campos, O, Agarwal, R, Allaire, M, Orville, A.M, Sweet, R.M, Soares, A.S. | | Deposit date: | 2014-03-03 | | Release date: | 2014-04-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Acoustic transfer of protein crystals from agarose pedestals to micromeshes for high-throughput screening.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4PA1
 
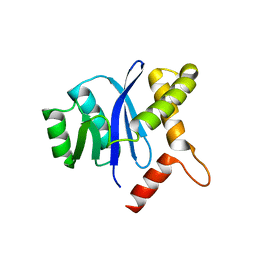 | |
4NCY
 
 | | In situ trypsin crystallized on a MiTeGen micromesh with imidazole ligand | | Descriptor: | 1,2-ETHANEDIOL, BENZAMIDINE, CALCIUM ION, ... | | Authors: | Yin, X, Scalia, A, Leroy, L, Cuttitta, C.M, Polizzo, G.M, Ericson, D.L, Roessler, C.G, Campos, O, Agarwal, R, Allaire, M, Orville, A.M, Jackimowicz, R, Ma, M.Y, Sweet, R.M, Soares, A.S. | | Deposit date: | 2013-10-25 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Hitting the target: fragment screening with acoustic in situ co-crystallization of proteins plus fragment libraries on pin-mounted data-collection micromeshes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2O4P
 
 | | Crystal Structure of HIV-1 Protease (Q7K) in Complex with Tipranavir | | Descriptor: | GLYCEROL, N-(3-{(1R)-1-[(6R)-4-HYDROXY-2-OXO-6-PHENETHYL-6-PROPYL-5,6-DIHYDRO-2H-PYRAN-3-YL]PROPYL}PHENYL)-5-(TRIFLUOROMETHYL)-2-PYRIDINESULFONAMIDE, protease | | Authors: | Kang, L.W, Armstrong, A.A, Muzammil, S, Jakalian, A, Bonneau, P.R, Schmelmer, V, Freire, E, Amzel, L.M. | | Deposit date: | 2006-12-04 | | Release date: | 2006-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unique thermodynamic response of tipranavir to human immunodeficiency virus type 1 protease drug resistance mutations.
J.Virol., 81, 2007
|
|
