8HF1
 
 | | DmDcr-2/R2D2/LoqsPD with 19bp-dsRNA in Trimer state | | Descriptor: | Dicer-2, isoform A, LD06392p, ... | | Authors: | Su, S, Wang, J, Ma, J. | | Deposit date: | 2022-11-09 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural mechanism of R2D2 and Loqs-PD synergistic modulation on DmDcr-2 oligomers.
Nat Commun, 14, 2023
|
|
7BWQ
 
 | | Structure of nonstructural protein Nsp9 from SARS-CoV-2 | | Descriptor: | Nsp9, SULFATE ION | | Authors: | Zhang, C, Chen, Y, Li, L, Su, D. | | Deposit date: | 2020-04-15 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.954 Å) | | Cite: | Structural basis for the multimerization of nonstructural protein nsp9 from SARS-CoV-2.
Mol Biomed, 1, 2020
|
|
3UQR
 
 | | Crystal structure of BACE1 with its inhibitor | | Descriptor: | Beta-secretase 1, METHYL (2S)-1-[(2R,5S,8S,12S,13S)-2,13-DIBENZYL-12-HYDROXY-3,5-DIMETHYL-15-(3-[METHYL(METHYLSULFONYL)AMINO]-5-{[(1R)-1-PHENYLETHYL]CARBAMOYL}PHENYL)-8-(2-METHYLPROPYL)-4,7,10,15-TETRAOXO-3,6,9,14-TETRAAZAPENTADECAN-1-OYL]PYRROLIDINE-2-CARBOXYLATE | | Authors: | Chen, T.T, Chen, W.Y, Li, L, Xu, Y.C. | | Deposit date: | 2011-11-21 | | Release date: | 2012-11-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.056 Å) | | Cite: | Cyanobacterial Peptides as a Prototype for the Design of Potent beta-Secretase Inhibitors and the Development of Selective Chemical Probes for Other Aspartic Proteases
J.Med.Chem., 55, 2012
|
|
7W0D
 
 | | Dicer2-LoqsPD-dsRNA complex at mid-translocation state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dicer-2, isoform A, ... | | Authors: | Su, S, Wang, J, Wang, H.W, Ma, J. | | Deposit date: | 2021-11-18 | | Release date: | 2022-04-27 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Structural insights into dsRNA processing by Drosophila Dicer-2-Loqs-PD.
Nature, 607, 2022
|
|
7W0C
 
 | | Dicer2-Loqs-PD-dsRNA complex at early-translocation state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dicer-2, isoform A, ... | | Authors: | Su, S, Wang, J, Wang, H.W, Ma, J. | | Deposit date: | 2021-11-18 | | Release date: | 2022-04-27 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Structural insights into dsRNA processing by Drosophila Dicer-2-Loqs-PD.
Nature, 607, 2022
|
|
7W0E
 
 | | dmDicer2-LoqsPD-dsRNA Active-dicing status | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dicer-2, isoform A, ... | | Authors: | Su, S, Wang, J, Wang, H.W, Ma, J. | | Deposit date: | 2021-11-18 | | Release date: | 2022-04-27 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | Structural insights into dsRNA processing by Drosophila Dicer-2-Loqs-PD.
Nature, 607, 2022
|
|
7W0B
 
 | | Dicer2-LoqsPD complex at apo status | | Descriptor: | Dicer-2, isoform A, Loquacious, ... | | Authors: | Su, S, Wang, J, Wang, H.W, Ma, J. | | Deposit date: | 2021-11-18 | | Release date: | 2022-04-27 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structural insights into dsRNA processing by Drosophila Dicer-2-Loqs-PD.
Nature, 607, 2022
|
|
7W0F
 
 | | dmDicer2-LoqsPD-dsRNA Post-dicing status | | Descriptor: | Dicer-2, isoform A, Loquacious, ... | | Authors: | Su, S, Wang, J, Wang, H.W, Ma, J. | | Deposit date: | 2021-11-18 | | Release date: | 2022-04-27 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.55 Å) | | Cite: | Structural insights into dsRNA processing by Drosophila Dicer-2-Loqs-PD.
Nature, 607, 2022
|
|
7W0A
 
 | | dmDicer2-LoqsPD-dsRNA Dimer status | | Descriptor: | Dicer-2, isoform A, Loquacious, ... | | Authors: | Su, S, Wang, J, Wang, H.W, Ma, J. | | Deposit date: | 2021-11-18 | | Release date: | 2022-04-27 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structural insights into dsRNA processing by Drosophila Dicer-2-Loqs-PD.
Nature, 607, 2022
|
|
5CDK
 
 | | Apical domain of chloroplast chaperonin 60b1 | | Descriptor: | Chaperonin 60B1 | | Authors: | Zhang, S, Yu, F, Liu, C, Gao, F. | | Deposit date: | 2015-07-04 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Functional Partition of Cpn60 alpha and Cpn60 beta Subunits in Substrate Recognition and Cooperation with Co-chaperonins
Mol Plant, 9, 2016
|
|
5CDJ
 
 | | apical domain of chloroplast chaperonin 60a | | Descriptor: | RuBisCO large subunit-binding protein subunit alpha, chloroplastic | | Authors: | Zhang, S, Yu, F, Liu, C, Gao, F. | | Deposit date: | 2015-07-04 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Functional Partition of Cpn60 alpha and Cpn60 beta Subunits in Substrate Recognition and Cooperation with Co-chaperonins
Mol Plant, 9, 2016
|
|
5CIO
 
 | | Crystal structure of PqqF | | Descriptor: | ZINC ION, pyrroloquinoline quinone biosynthesis protein PqqF | | Authors: | Wei, Q, Xu, D, Ran, T, Wang, W. | | Deposit date: | 2015-07-13 | | Release date: | 2016-06-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure and Function of PqqF Protein in the Pyrroloquinoline Quinone Biosynthetic Pathway
J.Biol.Chem., 291, 2016
|
|
5UJX
 
 | | Crystal structure of DHFR in 20% Isopropanol | | Descriptor: | CALCIUM ION, CHLORIDE ION, Dihydrofolate reductase, ... | | Authors: | Cuneo, M.J, Agarwal, P.K. | | Deposit date: | 2017-01-19 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modulating Enzyme Activity by Altering Protein Dynamics with Solvent.
Biochemistry, 57, 2018
|
|
7W8S
 
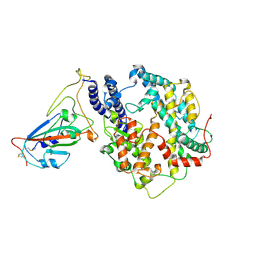 | | Structure of SARS-CoV-2 spike receptor-binding domain Y453F mutation complexed with American mink ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Su, C, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-12-08 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Molecular Basis of Mink ACE2 Binding to SARS-CoV-2 and Its Mink-Derived Variants.
J.Virol., 96, 2022
|
|
5EAJ
 
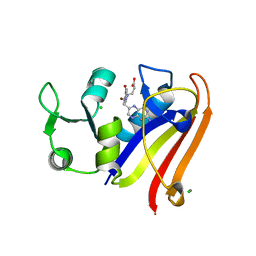 | | Crystal structure of DHFR in 0% Isopropanol | | Descriptor: | CALCIUM ION, CHLORIDE ION, Dihydrofolate reductase, ... | | Authors: | Cuneo, M.J, Agarwal, P.K. | | Deposit date: | 2015-10-16 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Modulating Enzyme Activity by Altering Protein Dynamics with Solvent.
Biochemistry, 57, 2018
|
|
7WA1
 
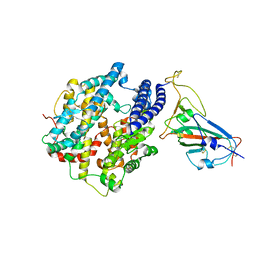 | |
6J66
 
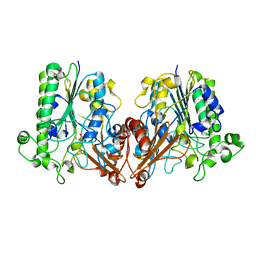 | | Chondroitin sulfate/dermatan sulfate endolytic 4-O-sulfatase | | Descriptor: | CALCIUM ION, Chondroitin sulfate/dermatan sulfate 4-O-endosulfatase protein | | Authors: | Gu, L, Li, F, Su, T, Wang, S. | | Deposit date: | 2019-01-14 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Comparative Study of Two Chondroitin Sulfate/Dermatan Sulfate 4-O-Sulfatases With High Identity.
Front Microbiol, 10, 2019
|
|
5XKC
 
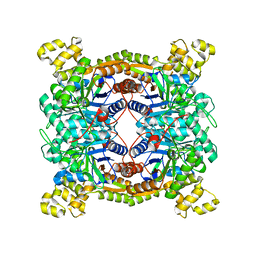 | | Crystal structure of dibenzothiophene sulfone monooxygenase BdsA at 2.2 angstrome | | Descriptor: | Dibenzothiophene desulfurization enzyme A | | Authors: | Gu, L, Su, T, Liu, S, Su, J. | | Deposit date: | 2017-05-07 | | Release date: | 2018-05-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.209 Å) | | Cite: | Structural and Biochemical Characterization of BdsA fromBacillus subtilisWU-S2B, a Key Enzyme in the "4S" Desulfurization Pathway.
Front Microbiol, 9, 2018
|
|
7FCP
 
 | |
7FCQ
 
 | |
5XKD
 
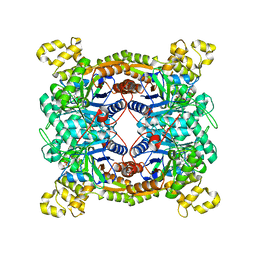 | | Crystal structure of dibenzothiophene sulfone monooxygenase BdsA in complex with FMN at 2.4 angstrom | | Descriptor: | Dibenzothiophene desulfurization enzyme A, FLAVIN MONONUCLEOTIDE | | Authors: | Gu, L, Su, T, Liu, S, Su, J. | | Deposit date: | 2017-05-07 | | Release date: | 2018-05-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Structural and Biochemical Characterization of BdsA fromBacillus subtilisWU-S2B, a Key Enzyme in the "4S" Desulfurization Pathway.
Front Microbiol, 9, 2018
|
|
7XR2
 
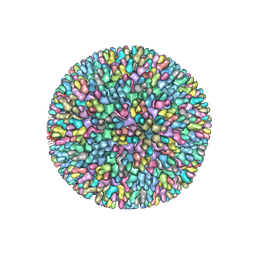 | |
7XR3
 
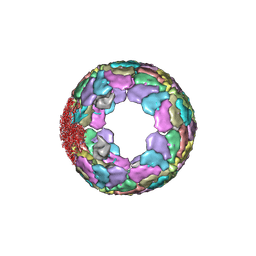 | |
2LKH
 
 | | WSA minor conformation | | Descriptor: | Acetylcholine receptor | | Authors: | Xu, Y, Mowrey, D, Cui, T, Perez-Aguilar, J, Saven, J.G, Eckenhoff, R, Tang, P. | | Deposit date: | 2011-10-11 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure and dynamics of a designed water-soluble transmembrane domain of nicotinic acetylcholine receptor.
Biochim.Biophys.Acta, 1818, 2011
|
|
7DWN
 
 | | Crystal structure of Vibrio fischeri DarR in complex with DNA reveals the transcriptional activation mechanism of LTTR family members | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Predicted DNA-binding transcriptional regulator | | Authors: | Wang, W.W, Wu, H, He, J.H, Yu, F. | | Deposit date: | 2021-01-17 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Crystal structure details of Vibrio fischeri DarR and mutant DarR-M202I from LTTR family reveals their activation mechanism.
Int.J.Biol.Macromol., 183, 2021
|
|
