6RXQ
 
 | | Crystal structure of CobB Ac2 (A76G,I131C,V162A) in complex with H4K16Cr-2'OH-ADPr peptide intermediate after soaking | | Descriptor: | Histone H4, NAD-dependent protein deacylase, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{R},4~{R},5~{S})-4-[(~{E})-but-2-enoxy]-3,5-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Spinck, M, Gasper, R, Neumann, H. | | Deposit date: | 2019-06-08 | | Release date: | 2020-04-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6RXM
 
 | | Crystal structure of CobB Ac2 (A76G, I131C, V162G) in complex with H4K16-Acetyl peptide | | Descriptor: | Histone H4, NAD-dependent protein deacylase, ZINC ION | | Authors: | Spinck, M, Gasper, R, Neumann, H. | | Deposit date: | 2019-06-08 | | Release date: | 2020-04-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6RXJ
 
 | | Crystal structure of CobB wt in complex with H4K16-Acetyl peptide | | Descriptor: | Histone H4, NAD-dependent protein deacylase, ZINC ION | | Authors: | Spinck, M, Gasper, R, Neumann, H. | | Deposit date: | 2019-06-08 | | Release date: | 2020-04-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7O2I
 
 | | METTL3-METTL14 heterodimer bound to the SAM competitive small molecule inhibitor STM2457 | | Descriptor: | DIMETHYL SULFOXIDE, N6-adenosine-methyltransferase catalytic subunit, N6-adenosine-methyltransferase non-catalytic subunit, ... | | Authors: | Pilka, E.S, Blackaby, W, Hardick, D, Harper, C, Hewstone, D, Ridgill, M, Rotty, B, Rausch, O. | | Deposit date: | 2021-03-30 | | Release date: | 2021-04-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia.
Nature, 593, 2021
|
|
4V0B
 
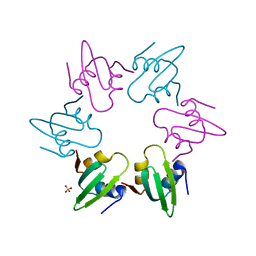 | | Escherichia coli FtsH hexameric N-domain | | Descriptor: | ATP-DEPENDENT ZINC METALLOPROTEASE FTSH, SULFATE ION | | Authors: | Serek-Heuberger, J, Martin, J, Lupas, A.N, Hartmann, M.D. | | Deposit date: | 2014-09-13 | | Release date: | 2015-01-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure and Evolution of N-Domains in Aaa Metalloproteases.
J.Mol.Biol., 427, 2015
|
|
5LW7
 
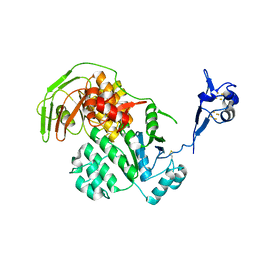 | | S. solfataricus ABCE1 post-splitting state | | Descriptor: | ABC transporter ATP-binding protein, IRON/SULFUR CLUSTER | | Authors: | Heuer, A, Gerovac, M, Beckmann, R, Tampe, R. | | Deposit date: | 2016-09-15 | | Release date: | 2016-11-16 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Structure of the ribosome post-recycling complex probed by chemical cross-linking and mass spectrometry.
Nat Commun, 7, 2016
|
|
5N65
 
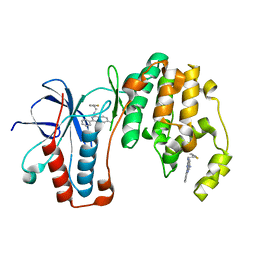 | | Crystal Structure of p38alpha in Complex with Lipid Pocket Ligand 9h | | Descriptor: | 2-phenyl-~{N}4-(2-thiophen-2-ylethyl)quinazoline-4,7-diamine, Mitogen-activated protein kinase 14 | | Authors: | Buehrmann, M, Mueller, M.P, Wiedemann, B, Rauh, D. | | Deposit date: | 2017-02-14 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design, synthesis and crystallization of 2-arylquinazolines as lipid pocket ligands of p38 alpha MAPK.
PLoS ONE, 12, 2017
|
|
5N64
 
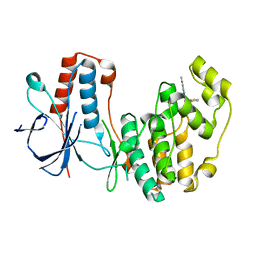 | | Crystal Structure of p38alpha in Complex with Lipid Pocket Ligand 9g | | Descriptor: | 2-phenyl-~{N}4-(thiophen-2-ylmethyl)quinazoline-4,7-diamine, Mitogen-activated protein kinase 14 | | Authors: | Buehrmann, M, Mueller, M.P, Wiedemann, B, Rauh, D. | | Deposit date: | 2017-02-14 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design, synthesis and crystallization of 2-arylquinazolines as lipid pocket ligands of p38 alpha MAPK.
PLoS ONE, 12, 2017
|
|
5N68
 
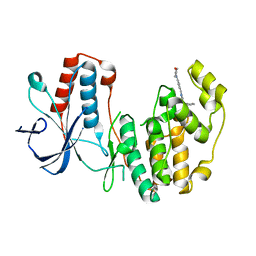 | | Crystal Structure of p38alpha in Complex with Lipid Pocket Ligand 9m | | Descriptor: | 2-(4-morpholin-4-ylphenyl)-~{N}4-(2-phenylethyl)quinazoline-4,7-diamine, Mitogen-activated protein kinase 14 | | Authors: | Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2017-02-14 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-based design, synthesis and crystallization of 2-arylquinazolines as lipid pocket ligands of p38 alpha MAPK.
PLoS ONE, 12, 2017
|
|
5LBY
 
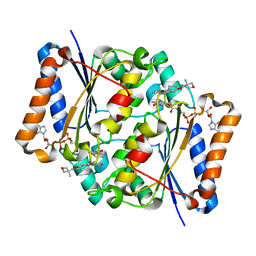 | | Structure of the human quinone reductase 2 (NQO2) in complex with crenolanib | | Descriptor: | 1-(2-{5-[(3-Methyloxetan-3-yl)methoxy]-1H-benzimidazol-1-yl}quinolin-8-yl)piperidin-4-amine, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Schneider, S, Medard, G, Kuester, B. | | Deposit date: | 2016-06-17 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The target landscape of clinical kinase drugs.
Science, 358, 2017
|
|
5N67
 
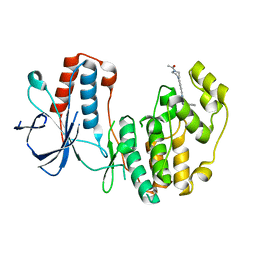 | |
5LBZ
 
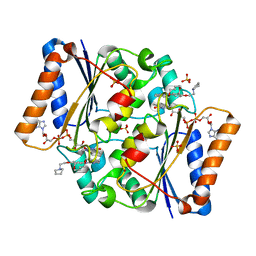 | | Structure of the human quinone reductase 2 (NQO2) in complex with pacritinib | | Descriptor: | 11-(2-pyrrolidin-1-yl-ethoxy)-14,19-dioxa-5,7,26-triaza-tetracyclo[19.3.1.1(2,6).1(8,12)]heptacosa-1(25),2(26),3,5,8,10,12(27),16,21,23-decaene, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Schneider, S, Medard, G, Kuster, B. | | Deposit date: | 2016-06-17 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The target landscape of clinical kinase drugs.
Science, 358, 2017
|
|
5LBW
 
 | | Structure of the human quinone reductase 2 (NQO2) in complex with volitinib | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ZINC ION, ... | | Authors: | Schneider, S, Medard, G, Kuester, B. | | Deposit date: | 2016-06-17 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The target landscape of clinical kinase drugs.
Science, 358, 2017
|
|
5M5A
 
 | | Crystal structure of MELK in complex with an inhibitor | | Descriptor: | CHLORIDE ION, K-252A, Maternal embryonic leucine zipper kinase, ... | | Authors: | Canevari, G, Re Depaolini, S, Casale, E, Felder, E, Kuster, B, Heinzlmeir, S. | | Deposit date: | 2016-10-21 | | Release date: | 2017-12-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The target landscape of clinical kinase drugs.
Science, 358, 2017
|
|
5MAH
 
 | | Crystal structure of MELK in complex with an inhibitor | | Descriptor: | CHLORIDE ION, GLYCEROL, Maternal embryonic leucine zipper kinase, ... | | Authors: | Canevari, G, Re Depaolini, S, Casale, E, Felder, E, Kuster, B, Heinzlmeir, S. | | Deposit date: | 2016-11-03 | | Release date: | 2017-12-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The target landscape of clinical kinase drugs.
Science, 358, 2017
|
|
5MAF
 
 | | Crystal structure of MELK in complex with an inhibitor | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Maternal embryonic leucine zipper kinase, ... | | Authors: | Canevari, G, Re Depaolini, S, Casale, E, Felder, E, Kuster, B, Heinzlmeir, S. | | Deposit date: | 2016-11-03 | | Release date: | 2017-12-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The target landscape of clinical kinase drugs.
Science, 358, 2017
|
|
5N66
 
 | | Crystal Structure of p38alpha in Complex with Lipid Pocket Ligand 9j | | Descriptor: | 1-(5-TERT-BUTYL-2-P-TOLYL-2H-PYRAZOL-3-YL)-3-[4-(2-MORPHOLIN-4-YL-ETHOXY)-NAPHTHALEN-1-YL]-UREA, Mitogen-activated protein kinase 14, ~{N}4-[[4-(cyclopropylmethyl)furan-2-yl]methyl]-2-phenyl-quinazoline-4,7-diamine | | Authors: | Buehrmann, M, Mueller, M.P, Wiedemann, B, Rauh, D. | | Deposit date: | 2017-02-14 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design, synthesis and crystallization of 2-arylquinazolines as lipid pocket ligands of p38 alpha MAPK.
PLoS ONE, 12, 2017
|
|
5MAI
 
 | | Crystal structure of MELK in complex with an inhibitor | | Descriptor: | 3-[(3~{Z})-3-[[[4-[(dimethylamino)methyl]phenyl]amino]-phenyl-methylidene]-2-oxidanylidene-1~{H}-indol-6-yl]-~{N}-ethyl-prop-2-ynamide, DIMETHYL SULFOXIDE, Maternal embryonic leucine zipper kinase | | Authors: | Canevari, G, Re Depaolini, S, Casale, E, Felder, E, Kuster, B, Heinzlmeir, S. | | Deposit date: | 2016-11-03 | | Release date: | 2017-12-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The target landscape of clinical kinase drugs.
Science, 358, 2017
|
|
5N63
 
 | | Crystal Structure of p38alpha in Complex with Lipid Pocket Ligand 9c | | Descriptor: | Mitogen-activated protein kinase 14, ~{N}4-[(4-fluorophenyl)methyl]-2-phenyl-quinazoline-4,7-diamine | | Authors: | Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2017-02-14 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design, synthesis and crystallization of 2-arylquinazolines as lipid pocket ligands of p38 alpha MAPK.
PLoS ONE, 12, 2017
|
|
7KDT
 
 | |
6YVV
 
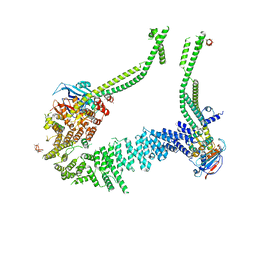 | | Condensin complex from S.cerevisiae ATP-free apo bridged state | | Descriptor: | Condensin complex subunit 1,Ycs4, Condensin complex subunit 2,Brn1, Structural maintenance of chromosomes protein 2,Structural maintenance of chromosomes protein 2, ... | | Authors: | Lee, B.-G, Cawood, C, Gutierrez-Escribano, P, Nakane, T, Merkel, F, Hassler, M, Haering, C.H, Aragon, L, Lowe, J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Cryo-EM structures of holo condensin reveal a subunit flip-flop mechanism.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6YVD
 
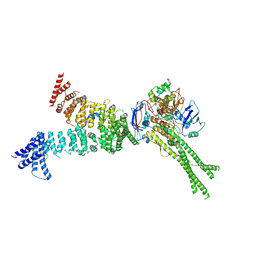 | | Head segment of the S.cerevisiae condensin holocomplex in presence of ATP | | Descriptor: | Condensin complex subunit 2, Condensin complex subunit 3, Structural maintenance of chromosomes protein 2, ... | | Authors: | Merkel, F, Haering, C.H, Hassler, M, Lee, B.G, Lowe, J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Cryo-EM structures of holo condensin reveal a subunit flip-flop mechanism.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6YVU
 
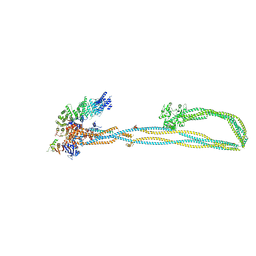 | | Condensin complex from S.cerevisiae ATP-free apo non-engaged state | | Descriptor: | Condensin complex subunit 1,Condensin complex subunit 1,Ycs4, Condensin complex subunit 2,Condensin complex subunit 2,Brn1, Structural maintenance of chromosomes protein 2,Structural maintenance of chromosomes protein 2,Smc2, ... | | Authors: | Lee, B.-G, Cawood, C, Gutierrez-Escribano, P, Nakane, T, Merkel, F, Hassler, M, Aragon, L, Haering, C.H, Lowe, J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-15 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Cryo-EM structures of holo condensin reveal a subunit flip-flop mechanism.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7OAO
 
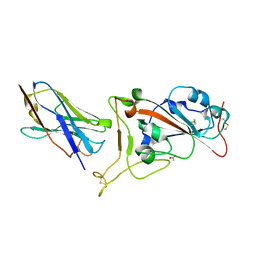 | | Nanobody C5 bound to RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, C5 nanobody, ... | | Authors: | Naismith, J.H, Mikolajek, H. | | Deposit date: | 2021-04-19 | | Release date: | 2021-08-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19.
Nat Commun, 12, 2021
|
|
7OAQ
 
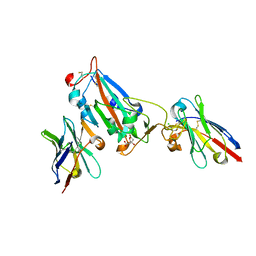 | | Nanobody H3 AND C1 bound to RBD with Kent mutation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Naismith, J.H, Mikolajek, H. | | Deposit date: | 2021-04-20 | | Release date: | 2021-08-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19.
Nat Commun, 12, 2021
|
|
