6C0S
 
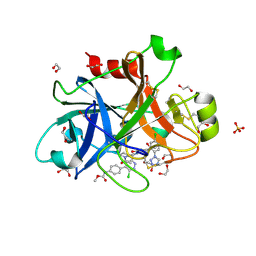 | |
5Q0G
 
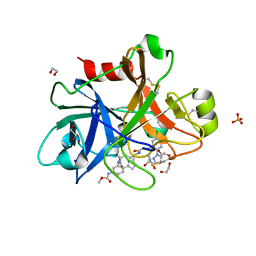 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR methyl [(3R,7S)-7-({(2E)-3-[5-chloro-2-(1H-tetrazol-1-yl)phenyl]prop-2-enoyl}amino)-3-ethyl-2-oxo-1,2,3,4,5,6,7,9-octahydro-11,8-(azeno)-1,9-benzodiazacyclotridecin-14-yl]carbamate | | Descriptor: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-05-01 | | Release date: | 2017-07-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Macrocyclic inhibitors of Factor XIa: Discovery of alkyl-substituted macrocyclic amide linkers with improved potency.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5WB6
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR methyl [(11S)-11-({(2E)-3-[5-chloro-2-(1H-tetrazol-1-yl)phenyl]prop-2-enoyl}amino)-6-fluoro-2-oxo-1,3,4,10,11,13-hexahydro-2H-5,9:15,12-di(azeno)-1,13-benzodiazacycloheptadecin-18-yl]carbamate | | Descriptor: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-06-28 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Macrocyclic factor XIa inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
4MQV
 
 | | Crystal complex of Rpa32c and Smarcal1 N-terminus | | Descriptor: | Replication protein A 32 kDa subunit, SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 | | Authors: | Xie, S, Qian, C.M. | | Deposit date: | 2013-09-16 | | Release date: | 2014-07-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of RPA32 bound to the N-terminus of SMARCAL1 redefines the binding interface between RPA32 and its interacting proteins
Febs J., 281, 2014
|
|
4MQ2
 
 | | The crystal structure of DYRK1a with a bound pyrido[2,3-d]pyrimidine inhibitor | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 1A, PENTAETHYLENE GLYCOL, SULFATE ION, ... | | Authors: | Lukacs, C.M, Janson, C.A, Garvie, C, Liang, L. | | Deposit date: | 2013-09-15 | | Release date: | 2013-12-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Pyrido[2,3-d]pyrimidines: Discovery and preliminary SAR of a novel series of DYRK1B and DYRK1A inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
5B5R
 
 | | Crystal structure of GSDMA3 | | Descriptor: | Gasdermin-A3 | | Authors: | Ding, J, Shao, F. | | Deposit date: | 2016-05-14 | | Release date: | 2016-06-15 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Pore-forming activity and structural autoinhibition of the gasdermin family.
Nature, 535, 2016
|
|
5TKT
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR METHYL ((12E,15S)-15-(((2E)-3-(5-CHLORO-2-(1H-TETRAZOL-1-YL)PHENYL)-2-PROPENOYL)AMINO)-9-OXO-8,17,19-TRIAZATRICYCLO[14.2.1.0~2,7~]NONADECA-1(18),2,4,6,12,16(19)-HEXAEN-5-YL)CARBAMATE | | Descriptor: | 1,2-ETHANEDIOL, Factor XIa (Light Chain), METHYL ((12E,15S)-15-(((2E)-3-(5-CHLORO-2-(1H-TETRAZOL-1-YL)PHENYL)-2-PROPENOYL)AMINO)-9-OXO-8,17,19-TRIAZATRICYCLO[14.2.1.0~2, ... | | Authors: | Sheriff, S. | | Deposit date: | 2016-10-07 | | Release date: | 2017-03-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure-Based Design of Macrocyclic Factor XIa Inhibitors: Discovery of the Macrocyclic Amide Linker.
J. Med. Chem., 60, 2017
|
|
5TKS
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR ((15S)-18-CHLORO- 15-(((2E)-3-(5-CHLORO-2-(1H-TETRAZOL-1-YL)PHENYL)-2- PROPENOYL)AMINO)-17,19-DIAZATRICYCLO[14.2.1.0~2,7~]NONADECA-1(18),2,4,6,16(19)-PENTAEN-5-YL)CARBAMATE | | Descriptor: | ((15S)-18-CHLORO- 15-(((2E)-3-(5-CHLORO-2-(1H-TETRAZOL-1-YL)PHENYL)-2- PROPENOYL)AMINO)-17,19-DIAZATRICYCLO[14.2.1.0~2,7~]NONADECA-1(18),2,4,6,16(19)-PENTAEN-5-YL)CARBAMATE, 1,2-ETHANEDIOL, Coagulation factor XI, ... | | Authors: | Sheriff, S. | | Deposit date: | 2016-10-07 | | Release date: | 2017-03-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Based Design of Macrocyclic Factor XIa Inhibitors: Discovery of the Macrocyclic Amide Linker.
J. Med. Chem., 60, 2017
|
|
5TKU
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR METHYL ((15S)-15-(((2E)-3-(5-CHLORO-2-(1H-TETRAZOL-1-YL)PHENYL)-2-PROPENOYL)AMINO)-9-OXO-8,17,19-TRIAZATRICYCLO[14.2.1.0~2,7~]NONADECA-1(18),2,4,6,16(19)-PENTAEN-5-YL)CARBAMATE | | Descriptor: | 1,2-ETHANEDIOL, Factor XIa (Light Chain), METHYL ((15S)-15-(((2E)-3-(5-CHLORO-2-(1H-TETRAZOL-1-YL)PHENYL)-2-PROPENOYL)AMINO)-9-OXO-8,17,19-TRIAZATRICYCLO[14.2.1.0~2,7~]N ONADECA-1(18),2,4,6,16(19)-PENTAEN-5-YL)CARBAMATE, ... | | Authors: | Sheriff, S. | | Deposit date: | 2016-10-07 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure-Based Design of Macrocyclic Factor XIa Inhibitors: Discovery of the Macrocyclic Amide Linker.
J. Med. Chem., 60, 2017
|
|
7YE9
 
 | | SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-05 | | Release date: | 2022-08-24 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
2XEM
 
 | | Induced-fit and allosteric effects upon polyene binding revealed by crystal structures of the Dynemicin thioesterase | | Descriptor: | (3E,5E,7E,9E,11E,13E)-pentadeca-3,5,7,9,11,13-hexaen-2-one, DYNE7 | | Authors: | Liew, C.W, Sharff, A, Kotaka, M, Kong, R, Bricogne, G, Liang, Z.X, Lescar, J. | | Deposit date: | 2010-05-17 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Induced-Fit Upon Ligand Binding Revealed by Crystal Structures of the Hot-Dog Fold Thioesterase in Dynemicin Biosynthesis.
J.Mol.Biol., 404, 2010
|
|
6LSA
 
 | | Complex structure of bovine herpesvirus 1 glycoprotein D and bovine nectin-1 IgV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein D, ... | | Authors: | Yue, D, Chen, Z.J, Yang, F.L, Ye, F, Lin, S, Cheng, Y.W, Wang, J.C, Chen, Z.M, Lin, X, Yang, J, Chen, H, Zhang, Z.L, You, Y, Sun, H.L, Wen, A, Wang, L.L, Zheng, Y, Cao, Y, Li, Y.H, Lu, G.W. | | Deposit date: | 2020-01-17 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Crystal structure of bovine herpesvirus 1 glycoprotein D bound to nectin-1 reveals the basis for its low-affinity binding to the receptor.
Sci Adv, 6, 2020
|
|
6LS9
 
 | | Crystal structure of bovine herpesvirus 1 glycoprotein D | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein D | | Authors: | Yue, D, Chen, Z.J, Yang, F.L, Ye, F, Lin, S, Cheng, Y.W, Wang, J.C, Chen, Z.M, Lin, X, Yang, J, Chen, H, Zhang, Z.L, You, Y, Sun, H.L, Wen, A, Wang, L.L, Zheng, Y, Cao, Y, Li, Y.H, Lu, G.W. | | Deposit date: | 2020-01-17 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Crystal structure of bovine herpesvirus 1 glycoprotein D bound to nectin-1 reveals the basis for its low-affinity binding to the receptor.
Sci Adv, 6, 2020
|
|
6U7N
 
 | | Crystal structure of neurotrimin (NTM) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Neurotrimin, ... | | Authors: | Machius, M, Venkannagari, H, Misra, A, Rudenko, G, Rush, S. | | Deposit date: | 2019-09-03 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.321 Å) | | Cite: | Highly Conserved Molecular Features in IgLONs Contrast Their Distinct Structural and Biological Outcomes.
J.Mol.Biol., 432, 2020
|
|
7OOR
 
 | | NaK C-DI mutant with Na+ and K+ | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ... | | Authors: | Minniberger, S, Plested, A.J.R. | | Deposit date: | 2021-05-28 | | Release date: | 2022-06-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Asymmetry and Ion Selectivity Properties of Bacterial Channel NaK Mutants Derived from Ionotropic Glutamate Receptors.
J.Mol.Biol., 435, 2023
|
|
7OPH
 
 | | NaK S-DI mutant with Na+ and K+ | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ... | | Authors: | Minniberger, S, Plested, A.J.R. | | Deposit date: | 2021-05-31 | | Release date: | 2022-06-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Asymmetry and Ion Selectivity Properties of Bacterial Channel NaK Mutants Derived from Ionotropic Glutamate Receptors.
J.Mol.Biol., 435, 2023
|
|
7OOU
 
 | | NaK C-DI mutant with Li+ and K+ | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Minniberger, S, Plested, A.J.R. | | Deposit date: | 2021-05-28 | | Release date: | 2022-06-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Asymmetry and Ion Selectivity Properties of Bacterial Channel NaK Mutants Derived from Ionotropic Glutamate Receptors.
J.Mol.Biol., 435, 2023
|
|
7OQ1
 
 | | NaK S-ELM mutant with Na+ and K+ | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Minniberger, S, Plested, A.J.R. | | Deposit date: | 2021-06-02 | | Release date: | 2022-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Asymmetry and Ion Selectivity Properties of Bacterial Channel NaK Mutants Derived from Ionotropic Glutamate Receptors.
J.Mol.Biol., 435, 2023
|
|
7P49
 
 | | HLA-E*01:03 in complex with Mtb14 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Walters, L.C, Gillespie, G.M. | | Deposit date: | 2021-07-10 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Primary and secondary functions of HLA-E are determined by stability and conformation of the peptide-bound complexes.
Cell Rep, 39, 2022
|
|
7OQ2
 
 | | NaK S-DI mutant soaked in Na+ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Minniberger, S, Plested, A.J.R. | | Deposit date: | 2021-06-02 | | Release date: | 2022-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Asymmetry and Ion Selectivity Properties of Bacterial Channel NaK Mutants Derived from Ionotropic Glutamate Receptors.
J.Mol.Biol., 435, 2023
|
|
7P4B
 
 | | HLA-E*01:03 in complex with IL9 | | Descriptor: | Beta-2-microglobulin, ESAT-6-like protein EsxH, GLYCEROL, ... | | Authors: | Walters, L.C, Gillespie, G.M. | | Deposit date: | 2021-07-10 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Primary and secondary functions of HLA-E are determined by stability and conformation of the peptide-bound complexes.
Cell Rep, 39, 2022
|
|
7PA0
 
 | | NaK C-DI F92A mutant with Rb+ and K+ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein, ... | | Authors: | Minniberger, S, Plested, A.J.R. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Asymmetry and Ion Selectivity Properties of Bacterial Channel NaK Mutants Derived from Ionotropic Glutamate Receptors.
J.Mol.Biol., 435, 2023
|
|
6R5F
 
 | | Crystal structure of RIP1 kinase in complex with DHP77 | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 1, [(5~{S})-5-[3,5-bis(fluoranyl)phenyl]pyrazolidin-1-yl]-[1-(5-methyl-1,3,4-oxadiazol-2-yl)piperidin-4-yl]methanone | | Authors: | Thorpe, J.H, Campobasso, N, Harris, P.A. | | Deposit date: | 2019-03-25 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Discovery and Lead-Optimization of 4,5-Dihydropyrazoles as Mono-Kinase Selective, Orally Bioavailable and Efficacious Inhibitors of Receptor Interacting Protein 1 (RIP1) Kinase.
J.Med.Chem., 62, 2019
|
|
8COI
 
 | | Human adenovirus-derived synthetic ADDobody binder | | Descriptor: | ADDobody | | Authors: | Buzas, D, Toelzer, C, Gupta, K, Berger-Schaffitzel, C, Berger, I. | | Deposit date: | 2023-02-28 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Engineering the ADDobody protein scaffold for generation of high-avidity ADDomer super-binders.
Structure, 32, 2024
|
|
4XXB
 
 | | Crystal structure of human MDM2-RPL11 | | Descriptor: | 60S ribosomal protein L11, BETA-MERCAPTOETHANOL, E3 ubiquitin-protein ligase Mdm2, ... | | Authors: | Zheng, J, Chen, Z. | | Deposit date: | 2015-01-30 | | Release date: | 2015-08-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human MDM2 complexed with RPL11 reveals the molecular basis of p53 activation
Genes Dev., 29, 2015
|
|
