3ZJ6
 
 | | Crystal of Raucaffricine Glucosidase in complex with inhibitor | | 分子名称: | (1R,2S,3S,4R,5R)-4-(cyclohexylmethylamino)-5-(hydroxymethyl)cyclopentane-1,2,3-triol, RAUCAFFRICINE-O-BETA-D-GLUCOSIDASE, SULFATE ION | | 著者 | Xia, L, Lin, H, Panjikar, S, Ruppert, M, Castiglia, A, Rajendran, C, Wang, M, Schuebel, H, Warzecha, H, Jaeger, V, Stoeckigt, J. | | 登録日 | 2013-01-17 | | 公開日 | 2014-01-29 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Ligand Structures of Synthetic Deoxa-Pyranosylamines with Raucaffricine and Strictosidine Glucosidases Provide Structural Insights Into Their Binding and Inhibitory Behaviours.
J.Enzyme.Inhib.Med.Chem., 30, 2015
|
|
3ZJ8
 
 | | Crystal structure of strictosidine glucosidase in complex with inhibitor-2 | | 分子名称: | (1R,2S,3S,4R,5R)-4-[(4-bromophenyl)methylamino]-5-(hydroxymethyl)cyclopentane-1,2,3-triol, STRICTOSIDINE-O-BETA-D-GLUCOSIDASE | | 著者 | Xia, L, Lin, H, Panjikar, S, Ruppert, M, Castiglia, A, Rajendran, C, Wang, M, Schuebel, H, Warzecha, H, Jaeger, V, Stoeckigt, J. | | 登録日 | 2013-01-17 | | 公開日 | 2014-02-05 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.01 Å) | | 主引用文献 | Ligand Structures of Synthetic Deoxa-Pyranosylamines with Raucaffricine and Strictosidine Glucosidases Provide Structural Insights Into Their Binding and Inhibitory Behaviours.
J.Enzyme.Inhib.Med.Chem., 30, 2015
|
|
4ATD
 
 | | Crystal structure of native Raucaffricine glucosidase | | 分子名称: | RAUCAFFRICINE-O-BETA-D-GLUCOSIDASE, SULFATE ION | | 著者 | Xia, L, Rajendran, C, Ruppert, M, Panjikar, S, Wang, M, Stoeckigt, J. | | 登録日 | 2012-05-05 | | 公開日 | 2013-01-16 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | High Speed X-Ray Analysis of Plant Enzymes at Room Temperature
Phytochemistry, 91, 2013
|
|
4ATL
 
 | | Crystal structure of Raucaffricine glucosidase in complex with Glucose | | 分子名称: | RAUCAFFRICINE-O-BETA-D-GLUCOSIDASE, beta-D-glucopyranose | | 著者 | Xia, L, Rajendran, C, Ruppert, M, Panjikar, S, Wang, M, Stoeckigt, J. | | 登録日 | 2012-05-08 | | 公開日 | 2013-01-30 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.52 Å) | | 主引用文献 | High Speed X-Ray Analysis of Plant Enzymes at Room Temperature
Phytochemistry, 91, 2013
|
|
4A3Y
 
 | | Crystal structure of Raucaffricine glucosidase from ajmaline biosynthesis pathway | | 分子名称: | GLYCEROL, RAUCAFFRICINE-O-BETA-D-GLUCOSIDASE, SULFATE ION | | 著者 | Xia, L, Ruppert, M, Wang, M, Panjikar, S, Barleben, L, Rajendran, C, Lin, H, Stoeckigt, J. | | 登録日 | 2011-10-06 | | 公開日 | 2012-08-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structures of Alkaloid Biosynthetic Glucosidases Decode Substrate Specificity.
Acs Chem.Biol., 7, 2012
|
|
3U57
 
 | | Structures of Alkaloid Biosynthetic Glucosidases Decode Substrate Specificity | | 分子名称: | (2beta,7beta,16S,17R,19E,21beta)-21-(beta-D-glucopyranosyloxy)-2,7-dihydro-7,17-cyclosarpagan-17-yl acetate, CHLORIDE ION, Raucaffricine-O-beta-D-glucosidase | | 著者 | Xia, L, Ruppert, M, Wang, M, Panjikar, S, Lin, H, Rajendran, C, Barleben, L, Stoeckigt, J. | | 登録日 | 2011-10-11 | | 公開日 | 2011-11-30 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.43 Å) | | 主引用文献 | Structures of alkaloid biosynthetic glucosidases decode substrate specificity.
Acs Chem.Biol., 7, 2012
|
|
3U5Y
 
 | | Structures of Alkaloid Biosynthetic Glucosidases Decode Substrate Specificity | | 分子名称: | CHLORIDE ION, Raucaffricine-O-beta-D-glucosidase, Secologanin | | 著者 | Xia, L, Ruppert, M, Wang, M, Panjikar, S, Lin, H, Rajendran, C, Barleben, L, Stoeckigt, J. | | 登録日 | 2011-10-11 | | 公開日 | 2011-11-30 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structures of alkaloid biosynthetic glucosidases decode substrate specificity.
Acs Chem.Biol., 7, 2012
|
|
3U5U
 
 | | Structures of Alkaloid Biosynthetic Glucosidases Decode Substrate Specificity | | 分子名称: | CHLORIDE ION, Raucaffricine-O-beta-D-glucosidase | | 著者 | Xia, L, Ruppert, M, Wang, M, Panjikar, S, Lin, H, Rajendran, C, Barleben, L, Stoeckigt, J. | | 登録日 | 2011-10-11 | | 公開日 | 2011-11-30 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structures of alkaloid biosynthetic glucosidases decode substrate specificity.
Acs Chem.Biol., 7, 2012
|
|
3ZJ7
 
 | | Crystal structure of strictosidine glucosidase in complex with inhibitor-1 | | 分子名称: | (1R,2S,3S,4R,5R)-4-(cyclohexylamino)-5-(hydroxymethyl)cyclopentane-1,2,3-triol, STRICTOSIDINE-O-BETA-D-GLUCOSIDASE | | 著者 | Xia, L, Lin, H, Panjikar, S, Ruppert, M, Castiglia, A, Rajendran, C, Wang, M, Schuebel, H, Warzecha, H, Jaeger, V, Stoeckigt, J. | | 登録日 | 2013-01-17 | | 公開日 | 2014-02-05 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Ligand Structures of Synthetic Deoxa-Pyranosylamines with Raucaffricine and Strictosidine Glucosidases Provide Structural Insights Into Their Binding and Inhibitory Behaviours.
J.Enzyme.Inhib.Med.Chem., 30, 2015
|
|
4EK7
 
 | | High speed X-ray analysis of plant enzymes at room temperature | | 分子名称: | CHLORIDE ION, Raucaffricine-O-beta-D-glucosidase, beta-D-glucopyranose | | 著者 | Xia, L, Rajendran, C, Ruppert, M, Panjikar, S, Wang, M, Stoeckigt, J. | | 登録日 | 2012-04-09 | | 公開日 | 2013-02-20 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | High speed X-ray analysis of plant enzymes at room temperature.
Phytochemistry, 2012
|
|
6LYU
 
 | | Structure of the BAM complex | | 分子名称: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | 著者 | Xiao, L, Huang, Y. | | 登録日 | 2020-02-15 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Structures of the beta-barrel assembly machine recognizing outer membrane protein substrates.
Faseb J., 35, 2021
|
|
6LYQ
 
 | | Structure of the BAM complex | | 分子名称: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | 著者 | Xiao, L, Huang, Y. | | 登録日 | 2020-02-15 | | 公開日 | 2021-01-06 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (3.19 Å) | | 主引用文献 | Structures of the beta-barrel assembly machine recognizing outer membrane protein substrates.
Faseb J., 35, 2021
|
|
6LYR
 
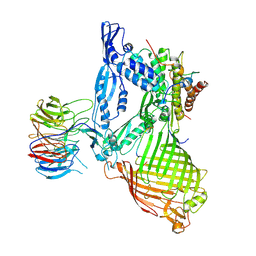 | | Structure of the BAM complex | | 分子名称: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | 著者 | Xiao, L, Huang, Y. | | 登録日 | 2020-02-15 | | 公開日 | 2021-01-06 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (3.28 Å) | | 主引用文献 | Structures of the beta-barrel assembly machine recognizing outer membrane protein substrates.
Faseb J., 35, 2021
|
|
6LM2
 
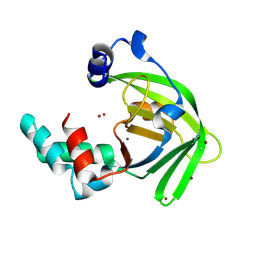 | |
6LPS
 
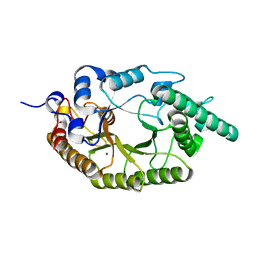 | |
6LYS
 
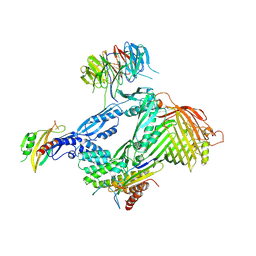 | | Structure of the BAM complex | | 分子名称: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | 著者 | Xiao, L, Huang, Y. | | 登録日 | 2020-02-15 | | 公開日 | 2021-01-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | Structures of the beta-barrel assembly machine recognizing outer membrane protein substrates.
Faseb J., 35, 2021
|
|
8JNX
 
 | |
8JN0
 
 | | N/F domain of alkaline amylase Amy703 | | 分子名称: | Alpha-amylase | | 著者 | Xiang, L, Zhang, G, Zhou, J. | | 登録日 | 2023-06-05 | | 公開日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.22826374 Å) | | 主引用文献 | Truncation of N-terminus domain of alkaline a-amylase to form a unique dimer leads to improved activity and stability and decreased calcium ion dependence
To Be Published
|
|
8JLV
 
 | |
8ERT
 
 | | NLRP3 PYD filament | | 分子名称: | NACHT, LRR and PYD domains-containing protein 3 | | 著者 | Wu, H, Xiao, L. | | 登録日 | 2022-10-12 | | 公開日 | 2022-12-14 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Cryo-EM structures of the active NLRP3 inflammasome disc.
Nature, 613, 2023
|
|
6M17
 
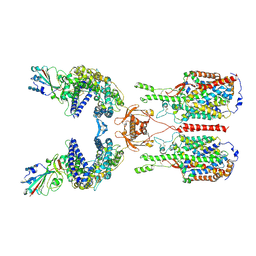 | | The 2019-nCoV RBD/ACE2-B0AT1 complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Yan, R.H, Zhang, Y.Y, Li, Y.N, Xia, L, Guo, Y.Y, Zhou, Q. | | 登録日 | 2020-02-24 | | 公開日 | 2020-03-11 | | 最終更新日 | 2021-03-10 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2.
Science, 367, 2020
|
|
6M1D
 
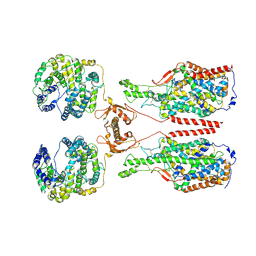 | | ACE2-B0AT1 complex, open conformation | | 分子名称: | Angiotensin-converting enzyme 2, Sodium-dependent neutral amino acid transporter B(0)AT1 | | 著者 | Yan, R.H, Zhang, Y.Y, Li, Y.N, Xia, L, Zhou, Q. | | 登録日 | 2020-02-25 | | 公開日 | 2020-03-11 | | 最終更新日 | 2020-04-08 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2.
Science, 367, 2020
|
|
6M18
 
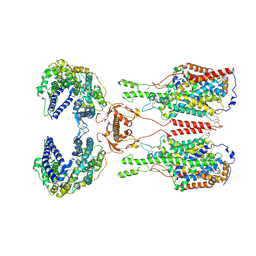 | | ACE2-B0AT1 complex | | 分子名称: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Yan, R.H, Zhang, Y.Y, Li, Y.N, Xia, L, Zhou, Q. | | 登録日 | 2020-02-25 | | 公開日 | 2020-03-11 | | 最終更新日 | 2020-11-04 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2.
Science, 367, 2020
|
|
7C2L
 
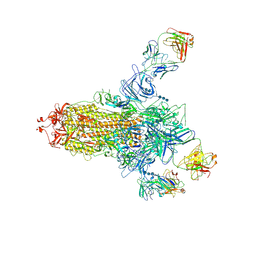 | | S protein of SARS-CoV-2 in complex bound with 4A8 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Yan, R.H, Zhang, Y.Y, Guo, Y.Y, Li, Y.N, Xia, L, Zhou, Q. | | 登録日 | 2020-05-08 | | 公開日 | 2020-07-01 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2.
Science, 369, 2020
|
|
7DX5
 
 | | S protein of SARS-CoV-2 bound with PD of ACE2 in the conformation 2 (1 up RBD and 1 PD bound) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Yan, R.H, Zhang, Y.Y, Li, Y.N, Ye, F.F, Guo, Y.Y, Xia, L, Zhong, X.Y, Chi, X.M, Zhou, Q. | | 登録日 | 2021-01-18 | | 公開日 | 2021-03-31 | | 最終更新日 | 2021-06-16 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural basis for the different states of the spike protein of SARS-CoV-2 in complex with ACE2.
Cell Res., 31, 2021
|
|
