2N0J
 
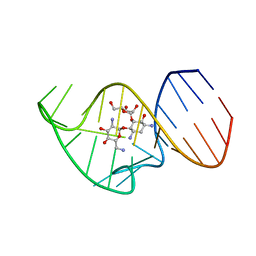 | | Solution NMR Structure of the 27 nucleotide engineered neomycin sensing riboswitch RNA-ribostamycin complex | | 分子名称: | RIBOSTAMYCIN, RNA_(27-MER) | | 著者 | Duchardt-Ferner, E, Gottstein-Schmidtke, S.R, Weigand, J.E, Ohlenschlaeger, O.E, Wurm, J, Hammann, C, Suess, B, Woehnert, J. | | 登録日 | 2015-03-09 | | 公開日 | 2016-02-03 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | What a Difference an OH Makes: Conformational Dynamics as the Basis for the Ligand Specificity of the Neomycin-Sensing Riboswitch.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
2KXM
 
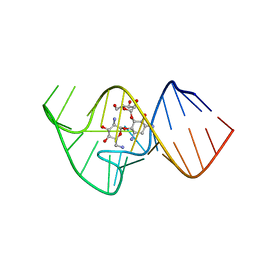 | | Solution NMR Structure of the 27 nucleotide engineered neomycin sensing riboswitch RNA-ribostmycin complex | | 分子名称: | RIBOSTAMYCIN, RNA (27-MER) | | 著者 | Duchardt-Ferner, E, Weigand, J.E, Ohlenschlager, O, Schmidtke, S.R, Suess, B, Wohnert, J. | | 登録日 | 2010-05-10 | | 公開日 | 2011-04-20 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Highly modular structure and ligand binding by conformational capture in a minimalistic riboswitch.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
6TQA
 
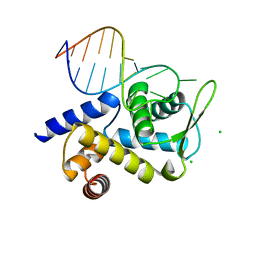 | | X-ray structure of Roquin ROQ domain in complex with a UCP3 CDE2 SL RNA motif | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, RNA (5'-R(P*GP*GP*UP*GP*CP*CP*UP*AP*AP*UP*AP*UP*UP*UP*AP*GP*GP*CP*AP*CP*(CCC))-3'), ... | | 著者 | Binas, O, Tants, J.-N, Peter, S.A, Janowski, R, Davydova, E, Braun, J, Niessing, D, Schwalbe, H, Weigand, J.E, Schlundt, A. | | 登録日 | 2019-12-16 | | 公開日 | 2020-05-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural basis for the recognition of transiently structured AU-rich elements by Roquin.
Nucleic Acids Res., 48, 2020
|
|
6TQB
 
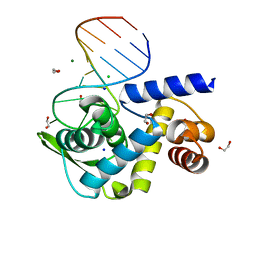 | | X-ray structure of Roquin ROQ domain in complex with a UCP3 CDE1 SL RNA motif | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Binas, O, Tants, J.-N, Peter, S.A, Janowski, R, Davydova, E, Braun, J, Niessing, D, Schwalbe, H, Weigand, J.E, Schlundt, A. | | 登録日 | 2019-12-16 | | 公開日 | 2020-05-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural basis for the recognition of transiently structured AU-rich elements by Roquin.
Nucleic Acids Res., 48, 2020
|
|
8CQ1
 
 | |
6XWW
 
 | |
6XXB
 
 | |
6XWJ
 
 | |
8BWT
 
 | |
8RHS
 
 | |
6XXA
 
 | |
2MXS
 
 | | Solution NMR-structure of the neomycin sensing riboswitch RNA bound to paromomycin | | 分子名称: | PAROMOMYCIN, RNA (27-MER) | | 著者 | Schmidtke, S, Duchardt-Ferner, E, Ohlenschlaeger, O, Gottstein, D, Wohnert, J. | | 登録日 | 2015-01-14 | | 公開日 | 2015-12-09 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | What a Difference an OH Makes: Conformational Dynamics as the Basis for the Ligand Specificity of the Neomycin-Sensing Riboswitch.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
