1FBK
 
 | |
1FBB
 
 | |
6CVM
 
 | | Atomic resolution cryo-EM structure of beta-galactosidase | | 分子名称: | 2-phenylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, MAGNESIUM ION, ... | | 著者 | Subramaniam, S, Bartesaghi, A, Banerjee, S, Zhu, X, Milne, J.L.S. | | 登録日 | 2018-03-28 | | 公開日 | 2018-05-30 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (1.9 Å) | | 主引用文献 | Atomic Resolution Cryo-EM Structure of beta-Galactosidase.
Structure, 26, 2018
|
|
4UQK
 
 | | Electron density map of GluA2em in complex with quisqualate and LY451646 | | 分子名称: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, GLUTAMATE RECEPTOR 2 | | 著者 | Meyerson, J.R, Kumar, J, Chittori, S, Rao, P, Pierson, J, Bartesaghi, A, Mayer, M.L, Subramaniam, S. | | 登録日 | 2014-06-24 | | 公開日 | 2014-08-13 | | 最終更新日 | 2017-08-02 | | 実験手法 | ELECTRON MICROSCOPY (16.4 Å) | | 主引用文献 | Structural Mechanism of Glutamate Receptor Activation and Desensitization
Nature, 514, 2014
|
|
6R1T
 
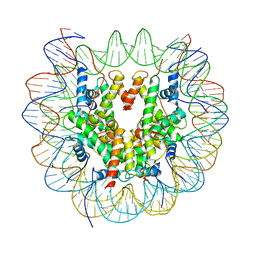 | | Structure of LSD2/NPAC-linker/nucleosome core particle complex: Class 1, free nuclesome | | 分子名称: | DNA (147-MER), HISTONE H2A, Histone H2A, ... | | 著者 | Marabelli, C, Pilotto, S, Chittori, S, Subramaniam, S, Mattevi, A. | | 登録日 | 2019-03-15 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (4.02 Å) | | 主引用文献 | A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex.
Cell Rep, 27, 2019
|
|
6R1U
 
 | | Structure of LSD2/NPAC-linker/nucleosome core particle complex: Class 2 | | 分子名称: | DNA (147-MER), FLAVIN-ADENINE DINUCLEOTIDE, Histone H2A, ... | | 著者 | Marabelli, C, Pilotto, S, Chittori, S, Subramaniam, S, Mattevi, A. | | 登録日 | 2019-03-15 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (4.36 Å) | | 主引用文献 | A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex.
Cell Rep, 27, 2019
|
|
6R25
 
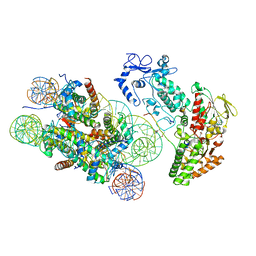 | | Structure of LSD2/NPAC-linker/nucleosome core particle complex: Class 3 | | 分子名称: | DNA (147-MER), FLAVIN-ADENINE DINUCLEOTIDE, H2B, ... | | 著者 | Marabelli, C, Pilotto, S, Chittori, S, Subramaniam, S, Mattevi, A. | | 登録日 | 2019-03-15 | | 公開日 | 2019-04-24 | | 実験手法 | ELECTRON MICROSCOPY (4.61 Å) | | 主引用文献 | A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex.
Cell Rep, 27, 2019
|
|
8VKM
 
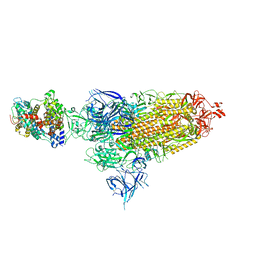 | | Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with mouse ACE2 (conformation 1) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Zhu, X, Mannar, D, Saville, J, Poloni, C, Bezeruk, A, Tidey, K, Ahmed, S, Tuttle, K, Vahdatihassani, F, Cholak, S, Cook, L, Steiner, T.S, Subramaniam, S. | | 登録日 | 2024-01-09 | | 公開日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (2.83 Å) | | 主引用文献 | SARS-CoV-2 XBB.1.5 Spike Protein: Altered Receptor Binding, Antibody Evasion, and Retention of T Cell Recognition
To Be Published
|
|
8VKK
 
 | | Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhu, X, Mannar, D, Saville, J, Poloni, C, Bezeruk, A, Tidey, K, Ahmed, S, Tuttle, K, Vahdatihassani, F, Cholak, S, Cook, L, Steiner, T.S, Subramaniam, S. | | 登録日 | 2024-01-09 | | 公開日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (2.81 Å) | | 主引用文献 | SARS-CoV-2 XBB.1.5 Spike Protein: Altered Receptor Binding, Antibody Evasion, and Retention of T Cell Recognition
To Be Published
|
|
8VKP
 
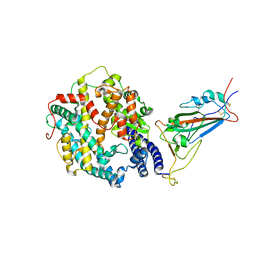 | | Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with human ACE2 (focused refinement of RBD and ACE2) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Zhu, X, Mannar, D, Saville, J, Poloni, C, Bezeruk, A, Tidey, K, Ahmed, S, Tuttle, K, Vahdatihassani, F, Cholak, S, Cook, L, Steiner, T.S, Subramaniam, S. | | 登録日 | 2024-01-09 | | 公開日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (2.77 Å) | | 主引用文献 | SARS-CoV-2 XBB.1.5 Spike Protein: Altered Receptor Binding, Antibody Evasion, and Retention of T Cell Recognition
To Be Published
|
|
8VKN
 
 | | Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with mouse ACE2 (focused refinement of RBD and mouse ACE2) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Zhu, X, Mannar, D, Saville, J, Poloni, C, Bezeruk, A, Tidey, K, Ahmed, S, Tuttle, K, Vahdatihassani, F, Cholak, S, Cook, L, Steiner, T.S, Subramaniam, S. | | 登録日 | 2024-01-09 | | 公開日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (2.93 Å) | | 主引用文献 | SARS-CoV-2 XBB.1.5 Spike Protein: Altered Receptor Binding, Antibody Evasion, and Retention of T Cell Recognition
To Be Published
|
|
8VKO
 
 | | Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with human ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Zhu, X, Mannar, D, Saville, J, Poloni, C, Bezeruk, A, Tidey, K, Ahmed, S, Tuttle, K, Vahdatihassani, F, Cholak, S, Cook, L, Steiner, T.S, Subramaniam, S. | | 登録日 | 2024-01-09 | | 公開日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (2.68 Å) | | 主引用文献 | SARS-CoV-2 XBB.1.5 Spike Protein: Altered Receptor Binding, Antibody Evasion, and Retention of T Cell Recognition
To Be Published
|
|
8VKL
 
 | | Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with mouse ACE2 (conformation 2) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Zhu, X, Mannar, D, Saville, J, Poloni, C, Bezeruk, A, Tidey, K, Ahmed, S, Tuttle, K, Vahdatihassani, F, Cholak, S, Cook, L, Steiner, T.S, Subramaniam, S. | | 登録日 | 2024-01-09 | | 公開日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (2.91 Å) | | 主引用文献 | SARS-CoV-2 XBB.1.5 Spike Protein: Altered Receptor Binding, Antibody Evasion, and Retention of T Cell Recognition
To Be Published
|
|
6EBM
 
 | | The voltage-activated Kv1.2-2.1 paddle chimera channel in lipid nanodiscs, transmembrane domain of subunit alpha | | 分子名称: | Potassium voltage-gated channel subfamily A member 2,Potassium voltage-gated channel subfamily B member 2 chimera | | 著者 | Matthies, D, Bae, C, Fox, T, Bartesaghi, A, Subramaniam, S, Swartz, K.J. | | 登録日 | 2018-08-06 | | 公開日 | 2018-08-22 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Single-particle cryo-EM structure of a voltage-activated potassium channel in lipid nanodiscs.
Elife, 7, 2018
|
|
6UAN
 
 | | B-Raf:14-3-3 complex | | 分子名称: | 14-3-3 zeta, Serine/threonine-protein kinase B-raf | | 著者 | Kondo, Y, Ognjenovic, J, Banerjee, S, Karandur, D, Merk, A, Kulhanek, K, Wong, K, Roose, J.P, Subramaniam, S, Kuriyan, J. | | 登録日 | 2019-09-11 | | 公開日 | 2019-09-25 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Cryo-EM structure of a dimeric B-Raf:14-3-3 complex reveals asymmetry in the active sites of B-Raf kinases.
Science, 366, 2019
|
|
5FTM
 
 | | Cryo-EM structure of human p97 bound to ATPgS (Conformation II) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Banerjee, S, Bartesaghi, A, Merk, A, Rao, P, Bulfer, S.L, Yan, Y, Green, N, Mroczkowski, B, Neitz, R.J, Wipf, P, Falconieri, V, Deshaies, R.J, Milne, J.L.S, Huryn, D, Arkin, M, Subramaniam, S. | | 登録日 | 2016-01-14 | | 公開日 | 2016-01-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | 2.3 A Resolution Cryo-Em Structure of Human P97 and Mechanism of Allosteric Inhibition
Science, 351, 2016
|
|
5FTK
 
 | | Cryo-EM structure of human p97 bound to ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, TRANSITIONAL ENDOPLASMIC RETICULUM ATPASE | | 著者 | Banerjee, S, Bartesaghi, A, Merk, A, Rao, P, Bulfer, S.L, Yan, Y, Green, N, Mroczkowski, B, Neitz, R.J, Wipf, P, Falconieri, V, Deshaies, R.J, Milne, J.L.S, Huryn, D, Arkin, M, Subramaniam, S. | | 登録日 | 2016-01-14 | | 公開日 | 2016-01-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (2.4 Å) | | 主引用文献 | 2.3 A Resolution Cryo-Em Structure of Human P97 and Mechanism of Allosteric Inhibition
Science, 351, 2016
|
|
5FTL
 
 | | Cryo-EM structure of human p97 bound to ATPgS (Conformation I) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, TRANSITIONAL ENDOPLASMIC RETICULUM ATPASE | | 著者 | Banerjee, S, Bartesaghi, A, Merk, A, Rao, P, Bulfer, S.L, Yan, Y, Green, N, Mroczkowski, B, Neitz, R.J, Wipf, P, Falconieri, V, Deshaies, R.J, Milne, J.L.S, Huryn, D, Arkin, M, Subramaniam, S. | | 登録日 | 2016-01-14 | | 公開日 | 2016-01-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | 2.3 A Resolution Cryo-Em Structure of Human P97 and Mechanism of Allosteric Inhibition
Science, 351, 2016
|
|
5FTJ
 
 | | Cryo-EM structure of human p97 bound to UPCDC30245 inhibitor | | 分子名称: | 1-(3-(5-FLUORO-1H-INDOL-2-YL)PHENYL)PIPERIDIN-4-YL)(2-(4-ISOPROPYL-PIPERAZIN1-YL)ETHYL)-CARBAMATE, ADENOSINE-5'-DIPHOSPHATE, TRANSITIONAL ENDOPLASMIC RETICULUM ATPASE | | 著者 | Banerjee, S, Bartesaghi, A, Merk, A, Rao, P, Bulfer, S.L, Yan, Y, Green, N, Mroczkowski, B, Neitz, R.J, Wipf, P, Falconieri, V, Deshaies, R.J, Milne, J.L.S, Huryn, D, Arkin, M, Subramaniam, S. | | 登録日 | 2016-01-14 | | 公開日 | 2016-01-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (2.3 Å) | | 主引用文献 | 2.3 A Resolution Cryo-Em Structure of Human P97 and Mechanism of Allosteric Inhibition
Science, 351, 2016
|
|
5FTN
 
 | | Cryo-EM structure of human p97 bound to ATPgS (Conformation III) | | 分子名称: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, TRANSITIONAL ENDOPLASMIC RETICULUM ATPASE | | 著者 | Banerjee, S, Bartesaghi, A, Merk, A, Rao, P, Bulfer, S.L, Yan, Y, Green, N, Mroczkowski, B, Neitz, R.J, Wipf, P, Falconieri, V, Deshaies, R.J, Milne, J.L.S, Huryn, D, Arkin, M, Subramaniam, S. | | 登録日 | 2016-01-14 | | 公開日 | 2016-01-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | 2.3 A Resolution Cryo-Em Structure of Human P97 and Mechanism of Allosteric Inhibition
Science, 351, 2016
|
|
2G8T
 
 | |
2G5V
 
 | |
6PWF
 
 | |
6PWE
 
 | |
4IFF
 
 | | Structural organization of FtsB, a transmembrane protein of the bacterial divisome | | 分子名称: | Fusion of phage phi29 Gp7 protein and Cell division protein FtsB, GLYCEROL | | 著者 | LaPointe, L.M, Taylor, K.C, Subramaniam, S, Khadria, A, Rayment, I, Senes, A. | | 登録日 | 2012-12-14 | | 公開日 | 2013-04-10 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural Organization of FtsB, a Transmembrane Protein of the Bacterial Divisome.
Biochemistry, 52, 2013
|
|
