6BS9
 
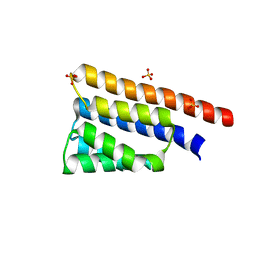 | | Stage III sporulation protein AB (SpoIIIAB) | | 分子名称: | SULFATE ION, Stage III sporulation protein AB | | 著者 | Strynadka, N.C.J, Zeytuni, N, Camp, A.H, Flanagan, K.A. | | 登録日 | 2017-12-01 | | 公開日 | 2018-01-17 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Structural characterization of SpoIIIAB sporulation-essential protein in Bacillus subtilis.
J. Struct. Biol., 202, 2018
|
|
6DCS
 
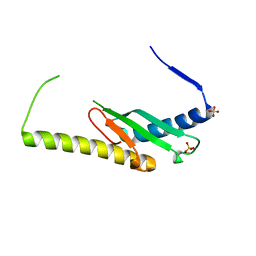 | | Stage III sporulation protein AF (SpoIIIAF) | | 分子名称: | SULFATE ION, Stage III sporulation protein AF | | 著者 | Strynadka, N.C.J, Zeytuni, N, Camp, A.H, Flanagan, K.A. | | 登録日 | 2018-05-08 | | 公開日 | 2018-07-18 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural and biochemical characterization of SpoIIIAF, a component of a sporulation-essential channel in Bacillus subtilis.
J. Struct. Biol., 204, 2018
|
|
6PEE
 
 | |
6ZTG
 
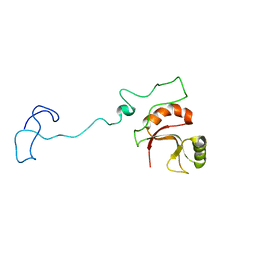 | | Spor protein DedD | | 分子名称: | Cell division protein DedD | | 著者 | Pazos, M, Peters, K, Boes, A, Safaei, Y, Kenward, C, Caveney, N.A, Laguri, C, Breukink, E, Strynadka, N.C.J, Simorre, J.P, Terrak, M, Vollmer, W. | | 登録日 | 2020-07-20 | | 公開日 | 2020-11-11 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | SPOR Proteins Are Required for Functionality of Class A Penicillin-Binding Proteins in Escherichia coli.
Mbio, 11, 2020
|
|
5TXI
 
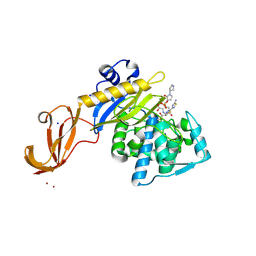 | | Crystal structure of wild-type S. aureus penicillin binding protein 4 (PBP4) in complex with ceftobiprole | | 分子名称: | (2R)-2-[(1R)-1-{[(2Z)-2-(5-amino-1,2,4-thiadiazol-3-yl)-2-(hydroxyimino)acetyl]amino}-2-oxoethyl]-5-({2-oxo-1-[(3R)-pyr rolidin-3-yl]-2,5-dihydro-1H-pyrrol-3-yl}methyl)-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | 著者 | Alexander, J.A.N, Strynadka, N.C.J. | | 登録日 | 2016-11-16 | | 公開日 | 2018-05-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural and kinetic analysis of penicillin-binding protein 4 (PBP4)-mediated antibiotic resistance inStaphylococcus aureus.
J. Biol. Chem., 2018
|
|
1KN9
 
 | |
7UZ2
 
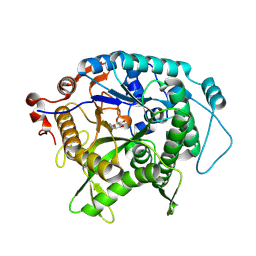 | | Structure of beta-glycosidase from Sulfolobus solfataricus in complex with C5a-fluoro-valienide. | | 分子名称: | (1R,2S,3R,4R)-5-fluoro-6-(hydroxymethyl)cyclohex-5-ene-1,2,3,4-tetrol, Beta-galactosidase | | 著者 | Danby, P.M, Jeong, A, Sim, L, Sweeney, R.P, Wardman, J.F, Geissner, A, Worrall, L.J, Strynadka, N.C.J, Withers, S.G. | | 登録日 | 2022-05-08 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Vinyl Halide-Modified Unsaturated Cyclitols are Mechanism-Based Glycosidase Inhibitors.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7UZ1
 
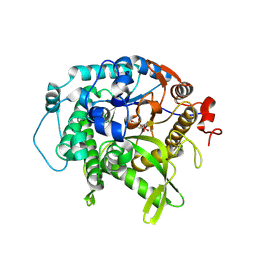 | | Structure of beta-glycosidase from Sulfolobus solfataricus in complex with C5a-bromo-valienide. | | 分子名称: | (1R,2S,3R,4R)-5-bromo-6-(hydroxymethyl)cyclohex-5-ene-1,2,3,4-tetrol, 1,2-ETHANEDIOL, Beta-galactosidase | | 著者 | Danby, P.M, Jeong, A, Sim, L, Sweeney, R.P, Wardman, J.F, Karimi, R, Geissner, A, Worrall, L.J, Strynadka, N.C.J, Withers, S.G. | | 登録日 | 2022-05-08 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Vinyl Halide-Modified Unsaturated Cyclitols are Mechanism-Based Glycosidase Inhibitors.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7TC5
 
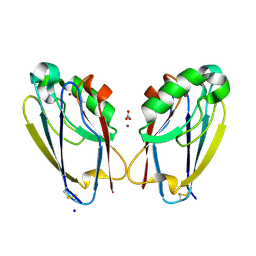 | | All Phe-Azurin variant - F15Y | | 分子名称: | Azurin, COPPER (II) ION, NITRATE ION, ... | | 著者 | Fedoretz-Maxwell, B.P, Worrall, L.J, Strynadka, N.C.J, Warren, J.J. | | 登録日 | 2021-12-22 | | 公開日 | 2022-06-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | The Impact of Second Coordination Sphere Methionine-Aromatic Interactions in Copper Proteins.
Inorg.Chem., 61, 2022
|
|
7TC6
 
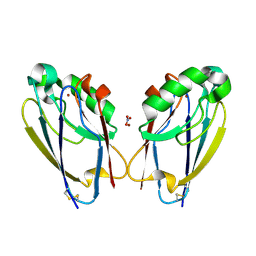 | | All Phe-Azurin variant - F15W | | 分子名称: | Azurin, COPPER (II) ION, NITRATE ION | | 著者 | Fedoretz-Maxwell, B.P, Worrall, L.J, Strynadka, N.C.J, Warren, J.J. | | 登録日 | 2021-12-22 | | 公開日 | 2022-06-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | The Impact of Second Coordination Sphere Methionine-Aromatic Interactions in Copper Proteins.
Inorg.Chem., 61, 2022
|
|
2DRJ
 
 | |
2F2H
 
 | | Structure of the YicI thiosugar Michaelis complex | | 分子名称: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, 4-NITROPHENYL 6-THIO-6-S-ALPHA-D-XYLOPYRANOSYL-BETA-D-GLUCOPYRANOSIDE, GLYCEROL, ... | | 著者 | Kim, Y.-W, Lovering, A.L, Strynadka, N.C.J, Withers, S.G. | | 登録日 | 2005-11-16 | | 公開日 | 2006-02-28 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Expanding the Thioglycoligase Strategy to the Synthesis of alpha-linked Thioglycosides Allows Structural Investigation of the Parent Enzyme/Substrate Complex
J.Am.Chem.Soc., 128, 2006
|
|
7LBU
 
 | | Crystal structure of the Propionibacterium acnes surface sialidase | | 分子名称: | ACETATE ION, Exo-alpha-sialidase, PHOSPHATE ION | | 著者 | Yu, A.C.Y, Volkers, G, Strynadka, N.C.J. | | 登録日 | 2021-01-08 | | 公開日 | 2021-12-08 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Crystal structure of the Propionibacterium acnes surface sialidase, a drug target for P. acnes-associated diseases.
Glycobiology, 32, 2022
|
|
7LBV
 
 | | Crystal structure of the Propionibacterium acnes surface sialidase in complex with Neu5Ac2en | | 分子名称: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, Exo-alpha-sialidase, PHOSPHATE ION | | 著者 | Yu, A.C.Y, Volkers, G, Strynadka, N.C.J. | | 登録日 | 2021-01-09 | | 公開日 | 2021-12-08 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of the Propionibacterium acnes surface sialidase, a drug target for P. acnes-associated diseases.
Glycobiology, 32, 2022
|
|
7LQ6
 
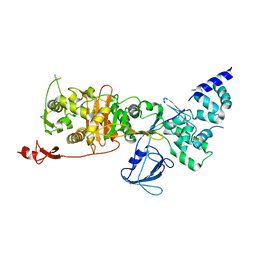 | | CryoEM structure of Escherichia coli PBP1b | | 分子名称: | Penicillin-binding protein 1B | | 著者 | Caveney, N.A, Workman, S.D, Yan, R, Atkinson, C.E, Yu, Z, Strynadka, N.C.J. | | 登録日 | 2021-02-13 | | 公開日 | 2021-05-26 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.28 Å) | | 主引用文献 | CryoEM structure of the antibacterial target PBP1b at 3.3 angstrom resolution.
Nat Commun, 12, 2021
|
|
6C39
 
 | |
6C3K
 
 | |
1Y5I
 
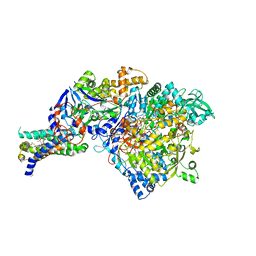 | | The crystal structure of the NarGHI mutant NarI-K86A | | 分子名称: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, FE3-S4 CLUSTER, ... | | 著者 | Bertero, M.G, Rothery, R.A, Boroumand, N, Palak, M, Blasco, F, Ginet, N, Weiner, J.H, Strynadka, N.C.J. | | 登録日 | 2004-12-02 | | 公開日 | 2005-03-08 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural and Biochemical Characterization of a Quinol Binding Site of Escherichia coli Nitrate Reductase A
J.Biol.Chem., 280, 2005
|
|
6PEM
 
 | | Focussed refinement of InvGN0N1:SpaPQR:PrgHK from Salmonella SPI-1 injectisome NC-base | | 分子名称: | Lipoprotein PrgK, Protein InvG, Protein PrgH, ... | | 著者 | Hu, J, Worrall, L.J, Strynadka, N.C.J. | | 登録日 | 2019-06-20 | | 公開日 | 2019-10-23 | | 最終更新日 | 2020-01-08 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | T3S injectisome needle complex structures in four distinct states reveal the basis of membrane coupling and assembly.
Nat Microbiol, 4, 2019
|
|
6Q15
 
 | | Structure of the Salmonella SPI-1 injectisome needle complex | | 分子名称: | Lipoprotein PrgK, Protein InvG, Protein PrgH, ... | | 著者 | Hu, J, Worrall, L.J, Strynadka, N.C.J. | | 登録日 | 2019-08-02 | | 公開日 | 2019-10-23 | | 最終更新日 | 2020-01-08 | | 実験手法 | ELECTRON MICROSCOPY (5.15 Å) | | 主引用文献 | T3S injectisome needle complex structures in four distinct states reveal the basis of membrane coupling and assembly.
Nat Microbiol, 4, 2019
|
|
6Q14
 
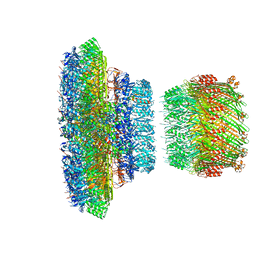 | | Structure of the Salmonella SPI-1 injectisome NC-base | | 分子名称: | Lipoprotein PrgK, Protein InvG, Protein PrgH, ... | | 著者 | Hu, J, Worrall, L.J, Strynadka, N.C.J. | | 登録日 | 2019-08-02 | | 公開日 | 2019-10-23 | | 最終更新日 | 2020-01-08 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | T3S injectisome needle complex structures in four distinct states reveal the basis of membrane coupling and assembly.
Nat Microbiol, 4, 2019
|
|
6Q16
 
 | | Focussed refinement of InvGN0N1:PrgHK:SpaPQR:PrgIJ from Salmonella SPI-1 injectisome NC-base | | 分子名称: | Lipoprotein PrgK, Protein InvG, Protein PrgH, ... | | 著者 | Hu, J, Worrall, L.J, Strynadka, N.C.J. | | 登録日 | 2019-08-02 | | 公開日 | 2019-10-23 | | 最終更新日 | 2020-01-15 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | T3S injectisome needle complex structures in four distinct states reveal the basis of membrane coupling and assembly.
Nat Microbiol, 4, 2019
|
|
5WD7
 
 | | Structure of a bacterial polysialyltransferase in complex with fondaparinux | | 分子名称: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-3,6-di-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-methyl 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranoside, SULFATE ION, SiaD | | 著者 | Worrall, L.J, Lizak, C, Strynadka, N.C.J. | | 登録日 | 2017-07-04 | | 公開日 | 2017-08-02 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | X-ray crystallographic structure of a bacterial polysialyltransferase provides insight into the biosynthesis of capsular polysialic acid.
Sci Rep, 7, 2017
|
|
6UEX
 
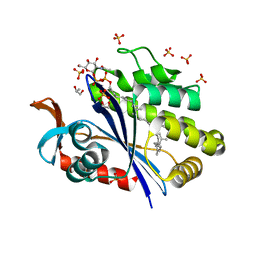 | | Crystal structure of S. aureus LcpA in complex with octaprenyl-pyrophosphate-GlcNAc | | 分子名称: | 2-(acetylamino)-2-deoxy-1-O-[(S)-hydroxy{[(S)-hydroxy{[(2Z,6Z,10Z,14Z,18Z,22Z,26Z)-3,7,11,15,19,23,27,31-octamethyldotriaconta-2,6,10,14,18,22,26,30-octaen-1-yl]oxy}phosphoryl]oxy}phosphoryl]-alpha-D-glucopyranose, GLYCEROL, Regulatory protein MsrR, ... | | 著者 | Li, F.K.K, Strynadka, N.C.J. | | 登録日 | 2019-09-23 | | 公開日 | 2020-01-29 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystallographic analysis ofStaphylococcus aureusLcpA, the primary wall teichoic acid ligase.
J.Biol.Chem., 295, 2020
|
|
6UF6
 
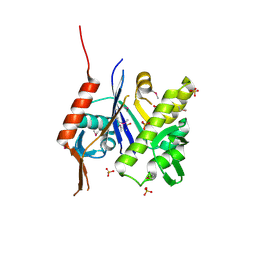 | | Crystal structure of B. subtilis TagU | | 分子名称: | GLYCEROL, Polyisoprenyl-teichoic acid--peptidoglycan teichoic acid transferase TagU, SULFATE ION | | 著者 | Li, F.K.K, Strynadka, N.C.J. | | 登録日 | 2019-09-23 | | 公開日 | 2020-01-29 | | 最終更新日 | 2020-03-11 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystallographic analysis ofStaphylococcus aureusLcpA, the primary wall teichoic acid ligase.
J.Biol.Chem., 295, 2020
|
|
