3DNY
 
 | |
1TRJ
 
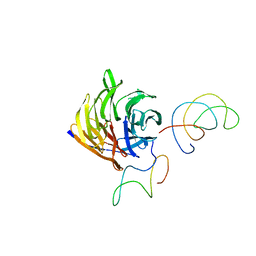 | | Homology Model of Yeast RACK1 Protein fitted into 11.7A cryo-EM map of Yeast 80S Ribosome | | 分子名称: | Guanine nucleotide-binding protein beta subunit-like protein, Helix 39 of 18S rRNA, Helix 40 of 18S rRNA | | 著者 | Sengupta, J, Nilsson, J, Gursky, R, Spahn, C.M, Nissen, P, Frank, J. | | 登録日 | 2004-06-21 | | 公開日 | 2004-09-28 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (11.7 Å) | | 主引用文献 | Identification of the versatile scaffold protein RACK1 on the eukaryotic ribosome by cryo-EM
Nat.Struct.Mol.Biol., 11, 2004
|
|
9K0Z
 
 | |
9K10
 
 | |
7Y41
 
 | |
7XAM
 
 | |
6IY7
 
 | |
6J45
 
 | |
6IZ7
 
 | |
6J0A
 
 | |
6IZI
 
 | |
6JR3
 
 | |
4V69
 
 | | Ternary complex-bound E.coli 70S ribosome. | | 分子名称: | 16S rRNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Villa, E, Sengupta, J, Trabuco, L.G, LeBarron, J, Baxter, W.T, Shaikh, T.R, Grassucci, R.A, Nissen, P, Ehrenberg, M, Schulten, K, Frank, J. | | 登録日 | 2008-12-11 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (6.7 Å) | | 主引用文献 | Ribosome-induced changes in elongation factor Tu conformation control GTP hydrolysis
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4V48
 
 | | Real space refined coordinates of the 30S and 50S subunits fitted into the low resolution cryo-EM map of the initiation-like state of E. coli 70S ribosome | | 分子名称: | 16S RIBOSOMAL RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | 著者 | Gao, H, Sengupta, J, Valle, M, Korostelev, A, Eswar, N, Stagg, S.M, Van Roey, P, Agrawal, R.K, Harvey, S.T, Sali, A, Chapman, M.S, Frank, J. | | 登録日 | 2003-05-06 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (11.5 Å) | | 主引用文献 | Study of the structural dynamics of the E. coli 70S ribosome using real space refinement
Cell(Cambridge,Mass.), 113, 2003
|
|
1ZN1
 
 | | Coordinates of RRF fitted into Cryo-EM map of the 70S post-termination complex | | 分子名称: | 30S ribosomal protein S12, Ribosome recycling factor, ribosomal 16S RNA, ... | | 著者 | Gao, N, Zavialov, A.V, Li, W, Sengupta, J, Valle, M, Gursky, R.P, Ehrenberg, M, Frank, J. | | 登録日 | 2005-05-11 | | 公開日 | 2005-06-14 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (14.1 Å) | | 主引用文献 | Mechanism for the disassembly of the posttermination complex inferred from cryo-EM studies.
Mol.Cell, 18, 2005
|
|
4V47
 
 | | Real space refined coordinates of the 30S and 50S subunits fitted into the low resolution cryo-EM map of the EF-G.GTP state of E. coli 70S ribosome | | 分子名称: | 16S RIBOSOMAL RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | 著者 | Gao, H, Sengupta, J, Valle, M, Korostelev, A, Eswar, N, Stagg, S.M, Van Roey, P, Agrawal, R.K, Harvey, S.T, Sali, A, Chapman, M.S, Frank, J. | | 登録日 | 2003-05-06 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (12.3 Å) | | 主引用文献 | Study of the structural dynamics of the E. coli 70S ribosome using real space refinement
Cell(Cambridge,Mass.), 113, 2003
|
|
1ZN0
 
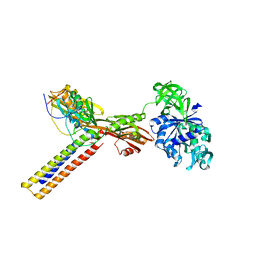 | | Coordinates of RRF and EF-G fitted into Cryo-EM map of the 50S subunit bound with both EF-G (GDPNP) and RRF | | 分子名称: | 16S RIBOSOMAL RNA, ELONGATION FACTOR G, Ribosome recycling factor | | 著者 | Gao, N, Zavialov, A.V, Li, W, Sengupta, J, Valle, M, Gursky, R.P, Ehrenberg, M, Frank, J. | | 登録日 | 2005-05-11 | | 公開日 | 2005-06-14 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (15.5 Å) | | 主引用文献 | Mechanism for the disassembly of the posttermination complex inferred from cryo-EM studies.
Mol.Cell, 18, 2005
|
|
8KAB
 
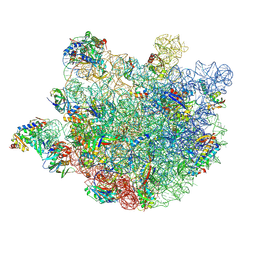 | | Mycobacterium smegmatis 50S ribosomal subunit-HflX complex | | 分子名称: | 23S rRNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | 著者 | Srinivasan, K, Banerjee, A, Sengupta, J. | | 登録日 | 2023-08-02 | | 公開日 | 2024-07-31 | | 最終更新日 | 2024-09-25 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Cryo-EM structures reveal the molecular mechanism of HflX-mediated erythromycin resistance in mycobacteria.
Structure, 32, 2024
|
|
8YP6
 
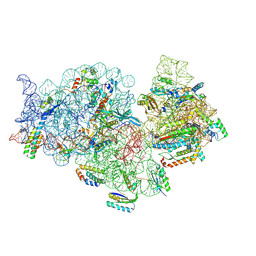 | |
8XZ3
 
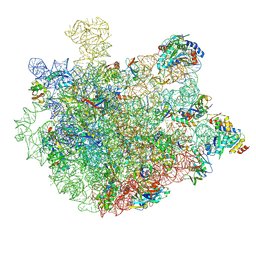 | | Mycobacterium smegmatis 50S ribosomal subunit with Erythromycin | | 分子名称: | 23S rRNA, 50S ribosomal protein L14, 50S ribosomal protein L19, ... | | 著者 | Srinivasan, K, Banerjee, A, Sengupta, J. | | 登録日 | 2024-01-20 | | 公開日 | 2024-07-31 | | 最終更新日 | 2024-09-25 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Cryo-EM structures reveal the molecular mechanism of HflX-mediated erythromycin resistance in mycobacteria.
Structure, 32, 2024
|
|
5XZC
 
 | |
3IYX
 
 | |
3IYY
 
 | |
1LU3
 
 | | Separate Fitting of the Anticodon Loop Region of tRNA (nucleotide 26-42) in the Low Resolution Cryo-EM Map of an EF-Tu Ternary Complex (GDP and Kirromycin) Bound to E. coli 70S Ribosome | | 分子名称: | PHENYLALANINE TRANSFER RNA | | 著者 | Valle, M, Sengupta, J, Swami, N.K, Grassucci, R.A, Burkhardt, N, Nierhaus, K.H, Agrawal, R.K, Frank, J. | | 登録日 | 2002-05-21 | | 公開日 | 2002-06-26 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (16.799999 Å) | | 主引用文献 | Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process.
EMBO J., 21, 2002
|
|
1LS2
 
 | | Fitting of EF-Tu and tRNA in the Low Resolution Cryo-EM Map of an EF-Tu Ternary Complex (GDP and Kirromycin) Bound to E. coli 70S Ribosome | | 分子名称: | Elongation Factor Tu, Phenylalanine transfer RNA | | 著者 | Valle, M, Sengupta, J, Swami, N.K, Grassucci, R.A, Burkhardt, N, Nierhaus, K.H, Agrawal, R.K, Frank, J. | | 登録日 | 2002-05-16 | | 公開日 | 2002-06-26 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (16.799999 Å) | | 主引用文献 | Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process.
EMBO J., 21, 2002
|
|
