4RQZ
 
 | |
4RR0
 
 | |
4RQY
 
 | |
4RR1
 
 | |
3OU0
 
 | | re-refined 3CS0 | | 分子名称: | Periplasmic serine endoprotease DegP, heptapeptide, pentapeptide | | 著者 | Sauer, R.T, Grant, R.A, Kim, S. | | 登録日 | 2010-09-14 | | 公開日 | 2011-01-19 | | 最終更新日 | 2012-02-22 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Covalent Linkage of Distinct Substrate Degrons Controls Assembly and Disassembly of DegP Proteolytic Cages.
Cell(Cambridge,Mass.), 145, 2011
|
|
6BQC
 
 | | Cyclopropane fatty acid synthase from E. coli | | 分子名称: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, CARBONATE ION, Cyclopropane-fatty-acyl-phospholipid synthase, ... | | 著者 | Hari, S.B, Grant, R.A, Sauer, R.T. | | 登録日 | 2017-11-27 | | 公開日 | 2018-07-04 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.073 Å) | | 主引用文献 | Structural and Functional Analysis of E. coli Cyclopropane Fatty Acid Synthase.
Structure, 26, 2018
|
|
1TWB
 
 | | SspB disulfide crosslinked to an ssrA degradation tag | | 分子名称: | Stringent starvation protein B homolog, ssrA peptide | | 著者 | Bolon, D.N, Grant, R.A, Baker, T.A, Sauer, R.T. | | 登録日 | 2004-06-30 | | 公開日 | 2004-11-16 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Nucleotide-Dependent Substrate Handoff from the SspB Adaptor to the AAA+ ClpXP Protease.
Mol.Cell, 16, 2004
|
|
1U9P
 
 | | Permuted single-chain Arc | | 分子名称: | pArc | | 著者 | Tabtiang, R.K, Cezairliyan, B.O, Grant, R.A, Cochrane, J.C, Sauer, R.T. | | 登録日 | 2004-08-10 | | 公開日 | 2005-02-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Consolidating critical binding determinants by noncyclic rearrangement of protein secondary structure
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
7UIY
 
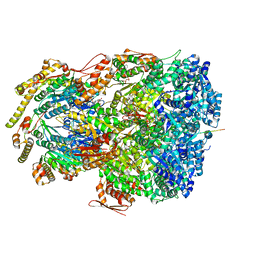 | | ClpAP complex bound to ClpS N-terminal extension, class IIIa | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | 著者 | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | 登録日 | 2022-03-29 | | 公開日 | 2022-10-26 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.22 Å) | | 主引用文献 | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UIW
 
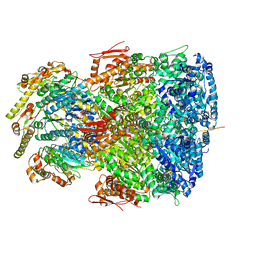 | | ClpAP complex bound to ClpS N-terminal extension, class IIb | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | 著者 | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | 登録日 | 2022-03-29 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.33 Å) | | 主引用文献 | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UJ0
 
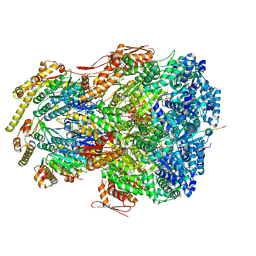 | | ClpAP complex bound to ClpS N-terminal extension, class IIIb | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | 著者 | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | 登録日 | 2022-03-29 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.26 Å) | | 主引用文献 | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UIZ
 
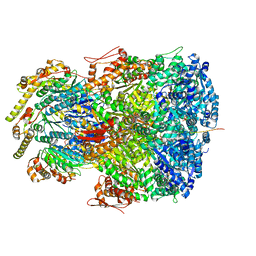 | | ClpAP complex bound to ClpS N-terminal extension, class IIc | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | 著者 | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | 登録日 | 2022-03-29 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.24 Å) | | 主引用文献 | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UIX
 
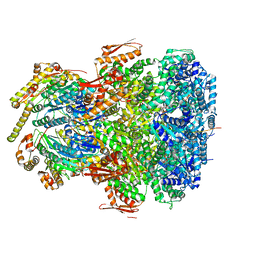 | | ClpAP complex bound to ClpS N-terminal extension, class I | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | 著者 | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | 登録日 | 2022-03-29 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.24 Å) | | 主引用文献 | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UIV
 
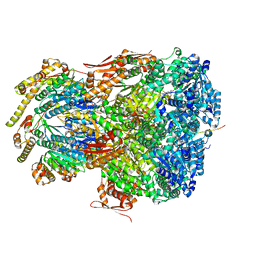 | | ClpAP complex bound to ClpS N-terminal extension, class IIa | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | 著者 | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | 登録日 | 2022-03-29 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.38 Å) | | 主引用文献 | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
1BDT
 
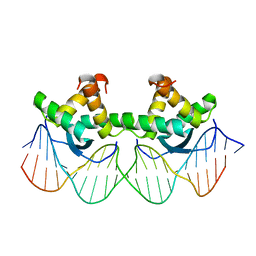 | | WILD TYPE GENE-REGULATING PROTEIN ARC/DNA COMPLEX | | 分子名称: | DNA (5'-D(*AP*AP*TP*GP*AP*TP*AP*GP*AP*AP*GP*CP*AP*CP*TP*CP*TP*AP*CP*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*GP*TP*AP*GP*AP*GP*TP*GP*CP*TP*TP*CP*TP*AP*TP*CP*AP*T)-3'), PROTEIN (GENE-REGULATING PROTEIN ARC) | | 著者 | Schilbach, J.F, Karzai, A.W, Raumann, B.E, Sauer, R.T. | | 登録日 | 1998-05-11 | | 公開日 | 1999-02-16 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Origins of DNA-binding specificity: role of protein contacts with the DNA backbone.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
7M1M
 
 | | Crystal structure of Pseudomonas aeruginosa ClpP1 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ATP-dependent Clp protease proteolytic subunit | | 著者 | Mawla, G.D, Grant, R.A, Baker, T.A, Sauer, R.T. | | 登録日 | 2021-03-13 | | 公開日 | 2021-03-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | ClpP1P2 peptidase activity promotes biofilm formation in Pseudomonas aeruginosa.
Mol.Microbiol., 115, 2021
|
|
7M1L
 
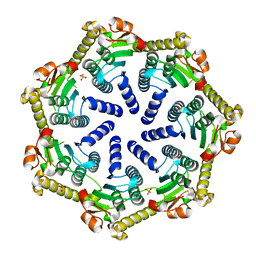 | | Crystal structure of Pseudomonas aeruginosa ClpP2 | | 分子名称: | ATP-dependent Clp protease proteolytic subunit, PHOSPHATE ION | | 著者 | Hall, B.M, Grant, R.A, Baker, T.A, Sauer, R.T. | | 登録日 | 2021-03-13 | | 公開日 | 2021-03-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | ClpP1P2 peptidase activity promotes biofilm formation in Pseudomonas aeruginosa.
Mol.Microbiol., 115, 2021
|
|
1YFN
 
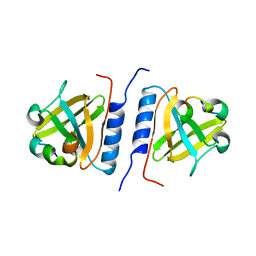 | | Versatile modes of peptide recognition by the AAA+ adaptor protein SspB- the crystal structure of a SspB-RseA complex | | 分子名称: | Sigma-E factor negative regulatory protein, Stringent starvation protein B | | 著者 | Levchenko, I, Grant, R.A, Flynn, J.M, Sauer, R.T, Baker, T.A. | | 登録日 | 2005-01-03 | | 公開日 | 2005-05-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Versatile modes of peptide recognition by the AAA+ adaptor protein SspB
Nat.Struct.Mol.Biol., 12, 2005
|
|
6WR2
 
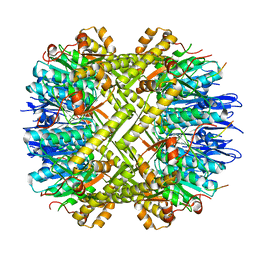 | |
6WRF
 
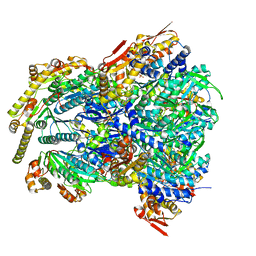 | | ClpX-ClpP complex bound to GFP-ssrA, recognition complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ATP-dependent Clp protease proteolytic subunit, ... | | 著者 | Fei, X, Sauer, R.T. | | 登録日 | 2020-04-29 | | 公開日 | 2020-11-04 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.14 Å) | | 主引用文献 | Structural basis of ClpXP recognition and unfolding of ssrA-tagged substrates.
Elife, 9, 2020
|
|
6WSG
 
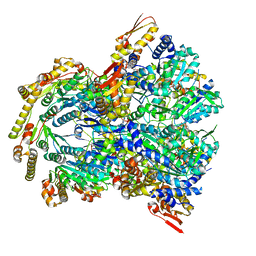 | |
5TXR
 
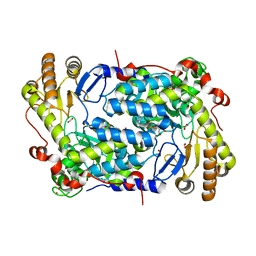 | | Structure of ALAS from S. cerevisiae non-covalently bound to PLP cofactor | | 分子名称: | 5-aminolevulinate synthase, mitochondrial, FORMIC ACID, ... | | 著者 | Brown, B.L, Grant, R.A, Kardon, J.R, Sauer, R.T, Baker, T.A. | | 登録日 | 2016-11-17 | | 公開日 | 2018-03-28 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of the Mitochondrial Aminolevulinic Acid Synthase, a Key Heme Biosynthetic Enzyme.
Structure, 26, 2018
|
|
5TXV
 
 | | HslU P21 cell with 4 hexamers | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent protease ATPase subunit HslU | | 著者 | Grant, R.A, Chen, J, Glynn, S.E, Sauer, R.T. | | 登録日 | 2016-11-17 | | 公開日 | 2017-03-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (7.086 Å) | | 主引用文献 | Covalently linked HslU hexamers support a probabilistic mechanism that links ATP hydrolysis to protein unfolding and translocation.
J. Biol. Chem., 292, 2017
|
|
5TXT
 
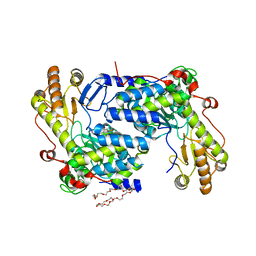 | | Structure of asymmetric apo/holo ALAS dimer from S. cerevisiae | | 分子名称: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, 5-aminolevulinate synthase, mitochondrial, ... | | 著者 | Brown, B.L, Grant, R.A, Kardon, J.R, Sauer, R.T, Baker, T.A. | | 登録日 | 2016-11-17 | | 公開日 | 2018-03-28 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure of the Mitochondrial Aminolevulinic Acid Synthase, a Key Heme Biosynthetic Enzyme.
Structure, 26, 2018
|
|
2HDD
 
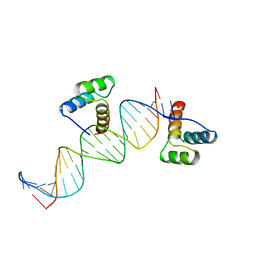 | | ENGRAILED HOMEODOMAIN Q50K VARIANT DNA COMPLEX | | 分子名称: | DNA (5'-D(*AP*TP*CP*CP*GP*GP*GP*GP*AP*TP*TP*AP*CP*AP*TP*GP*G P*CP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*TP*GP*CP*CP*AP*TP*GP*TP*AP*AP*TP*CP*CP*C P*CP*GP*GP*A)-3'), PROTEIN (ENGRAILED HOMEODOMAIN Q50K) | | 著者 | Tucker-Kellogg, L, Rould, M.A, Chambers, K.A, Ades, S.E, Sauer, R.T, Pabo, C.O. | | 登録日 | 1998-02-10 | | 公開日 | 1998-05-27 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Engrailed (Gln50-->Lys) homeodomain-DNA complex at 1.9 A resolution: structural basis for enhanced affinity and altered specificity.
Structure, 5, 1997
|
|
