4V8L
 
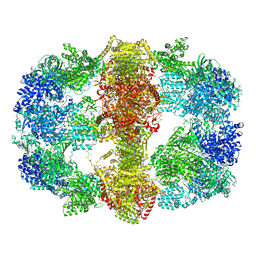 | |
2AGN
 
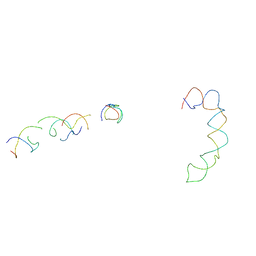 | | Fitting of hepatitis C virus internal ribosome entry site domains into the 15 A Cryo-EM map of a HCV IRES-80S ribosome (H. sapiens) complex | | 分子名称: | 6 nt A-RNA helix, HCV IRES DOMAIN II, HCV IRES IIIABC, ... | | 著者 | Boehringer, D, Thermann, R, Ostareck-Lederer, A, Lewis, J.D, Stark, H. | | 登録日 | 2005-07-27 | | 公開日 | 2006-07-25 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (15 Å) | | 主引用文献 | Structure of the hepatitis C Virus IRES bound to the human 80S ribosome: remodeling of the HCV IRES
Structure, 13, 2005
|
|
4ADV
 
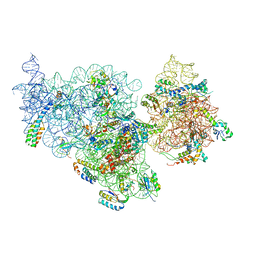 | | Structure of the E. coli methyltransferase KsgA bound to the E. coli 30S ribosomal subunit | | 分子名称: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | 著者 | Boehringer, D, O'Farrell, H.C, Rife, J.P, Ban, N. | | 登録日 | 2012-01-03 | | 公開日 | 2012-02-15 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (13.5 Å) | | 主引用文献 | Structural Insights Into Methyltransferase Ksga Function in 30S Ribosomal Subunit Biogenesis
J.Biol.Chem., 287, 2012
|
|
5LOQ
 
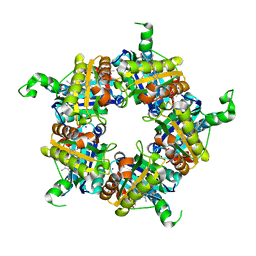 | | Structure of coproheme bound HemQ from Listeria monocytogenes | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, Putative heme-dependent peroxidase lmo2113, ... | | 著者 | Puehringer, D, Mlynek, G, Hofbauer, S, Djinovic-Carugo, K, Obinger, C. | | 登録日 | 2016-08-09 | | 公開日 | 2016-10-26 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Hydrogen peroxide-mediated conversion of coproheme to heme b by HemQ-lessons from the first crystal structure and kinetic studies.
FEBS J., 283, 2016
|
|
5K8Z
 
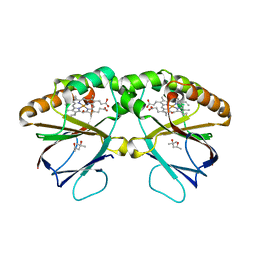 | | Crystal structure of dimeric chlorite dismutase from Cyanothece sp. PCC7425 (pH 8.5) | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Chlorite dismutase, ... | | 著者 | Puehringer, D, Schaffner, I, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | 登録日 | 2016-05-31 | | 公開日 | 2017-06-21 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Molecular Mechanism of Enzymatic Chlorite Detoxification: Insights from Structural and Kinetic Studies.
ACS Catal, 7, 2017
|
|
5K91
 
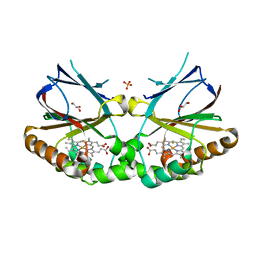 | | Crystal structure of dimeric chlorite dismutase from Cyanothece sp. PCC7425 in complex with fluoride | | 分子名称: | Chlorite dismutase, FLUORIDE ION, GLYCEROL, ... | | 著者 | Puehringer, D, Schaffner, I, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | 登録日 | 2016-05-31 | | 公開日 | 2017-06-21 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.18 Å) | | 主引用文献 | Molecular Mechanism of Enzymatic Chlorite Detoxification: Insights from Structural and Kinetic Studies.
ACS Catal, 7, 2017
|
|
5K90
 
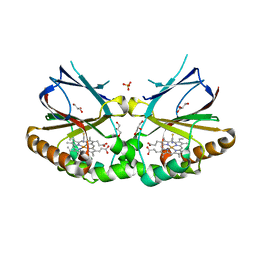 | | Crystal structure of dimeric chlorite dismutase from Cyanothece sp. PCC7425 in complex with isothiocyanate | | 分子名称: | Chlorite dismutase, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Puehringer, D, Schaffner, I, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | 登録日 | 2016-05-31 | | 公開日 | 2017-06-21 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.28 Å) | | 主引用文献 | Molecular Mechanism of Enzymatic Chlorite Detoxification: Insights from Structural and Kinetic Studies.
ACS Catal, 7, 2017
|
|
5MAU
 
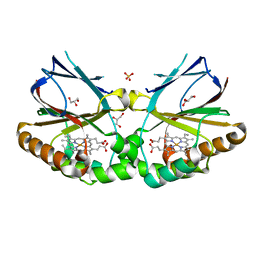 | | Crystal structure of dimeric chlorite dismutase from Cyanothece sp. PCC7425 (pH 6.5) | | 分子名称: | Chlorite dismutase, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Puehringer, D, Schaffner, I, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | 登録日 | 2016-11-04 | | 公開日 | 2017-12-20 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Molecular Mechanism of Enzymatic Chlorite Detoxification: Insights from Structural and Kinetic Studies.
ACS Catal, 7, 2017
|
|
5NKV
 
 | | Crystal structure of dimeric chlorite dismutase from Cyanothece sp. PCC7425 at pH 9.0 and 293 K. | | 分子名称: | CHLORIDE ION, Chlorite Dismutase, GLYCEROL, ... | | 著者 | Puehringer, D, Schaffner, I, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | 登録日 | 2017-04-03 | | 公開日 | 2018-01-31 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Molecular Mechanism of Enzymatic Chlorite Detoxification: Insights from Structural and Kinetic Studies.
ACS Catal, 7, 2017
|
|
5NKU
 
 | | Joint neutron/X-ray structure of dimeric chlorite dismutase from Cyanothece sp. PCC7425 | | 分子名称: | CHLORIDE ION, Chlorite Dismutase, GLYCEROL, ... | | 著者 | Puehringer, D, Schaffner, I, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | 登録日 | 2017-04-03 | | 公開日 | 2018-02-28 | | 最終更新日 | 2024-05-01 | | 実験手法 | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | 主引用文献 | Molecular Mechanism of Enzymatic Chlorite Detoxification: Insights from Structural and Kinetic Studies.
ACS Catal, 7, 2017
|
|
8CLT
 
 | |
8CLU
 
 | | Zearalenone lactonase from Rhodococcus erythropolis in complex with zearalactamenone | | 分子名称: | (4~{S})-4-methyl-16,18-bis(oxidanyl)-3-azabicyclo[12.4.0]octadeca-1(18),12,14,16-tetraene-2,8-dione, GLYCEROL, Zearalenone lactonase | | 著者 | Puehringer, D. | | 登録日 | 2023-02-17 | | 公開日 | 2024-02-21 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Bacterial Lactonases ZenA with Noncanonical Structural Features Hydrolyze the Mycotoxin Zearalenone.
Acs Catalysis, 14, 2024
|
|
8CLP
 
 | |
8CLN
 
 | | Zearalenone lactonase from Streptomyces coelicoflavus, SeMet derivative for SAD phasing | | 分子名称: | Hydrolase | | 著者 | Puehringer, D, Grishkovskaya, I, Mlynek, G, Kostan, J. | | 登録日 | 2023-02-17 | | 公開日 | 2024-02-21 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Bacterial Lactonases ZenA with Noncanonical Structural Features Hydrolyze the Mycotoxin Zearalenone.
Acs Catalysis, 14, 2024
|
|
8CLO
 
 | | Zearalenone lactonase from Streptomyces coelicoflavus | | 分子名称: | 1,2-ETHANEDIOL, Hydrolase | | 著者 | Puehringer, D, Grishkovskaya, I, Mlynek, G, Kostan, J. | | 登録日 | 2023-02-17 | | 公開日 | 2024-02-21 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Bacterial Lactonases ZenA with Noncanonical Structural Features Hydrolyze the Mycotoxin Zearalenone.
Acs Catalysis, 14, 2024
|
|
8CLV
 
 | |
8CLQ
 
 | |
8C0H
 
 | |
3CPO
 
 | | Crystal structure of ketosteroid isomerase D40N with bound 2-fluorophenol | | 分子名称: | 2-fluorophenol, Delta(5)-3-ketosteroid isomerase | | 著者 | Caaveiro, J.M.M, Pybus, B, Ringe, D, Petsko, G. | | 登録日 | 2008-03-31 | | 公開日 | 2008-09-23 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.24 Å) | | 主引用文献 | Testing geometrical discrimination within an enzyme active site: constrained hydrogen bonding in the ketosteroid isomerase oxyanion hole
J.Am.Chem.Soc., 130, 2008
|
|
4GCH
 
 | |
6GCH
 
 | | STRUCTURE OF CHYMOTRYPSIN-*TRIFLUOROMETHYL KETONE INHIBITOR COMPLEXES. COMPARISON OF SLOWLY AND RAPIDLY EQUILIBRATING INHIBITORS | | 分子名称: | 1,1,1-TRIFLUORO-3-ACETAMIDO-4-PHENYL BUTAN-2-ONE(N-ACETYL-L-PHENYLALANYL TRIFLUOROMETHYL KETONE), GAMMA-CHYMOTRYPSIN A | | 著者 | Brady, K, Wei, A, Ringe, D, Abeles, R.H. | | 登録日 | 1990-04-06 | | 公開日 | 1990-10-15 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of chymotrypsin-trifluoromethyl ketone inhibitor complexes: comparison of slowly and rapidly equilibrating inhibitors.
Biochemistry, 29, 1990
|
|
3LQS
 
 | | Complex Structure of D-Amino Acid Aminotransferase and 4-amino-4,5-dihydro-thiophenecarboxylic acid (ADTA) | | 分子名称: | 4-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]THIOPHENE-2-CARBOXYLIC ACID, ACETIC ACID, D-alanine aminotransferase | | 著者 | Lepore, B.W, Liu, D, Peng, Y, Fu, M, Yasuda, C, Manning, J.M, Silverman, R.B, Ringe, D. | | 登録日 | 2010-02-10 | | 公開日 | 2010-03-16 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Chiral discrimination among aminotransferases: inactivation by 4-amino-4,5-dihydrothiophenecarboxylic acid.
Biochemistry, 49, 2010
|
|
1XCV
 
 | |
2R2D
 
 | | Structure of a quorum-quenching lactonase (AiiB) from Agrobacterium tumefaciens | | 分子名称: | GLYCEROL, PHOSPHATE ION, ZINC ION, ... | | 著者 | Liu, D, Thomas, P.W, Momb, J, Hoang, Q, Petsko, G.A, Ringe, D, Fast, W. | | 登録日 | 2007-08-24 | | 公開日 | 2007-10-09 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structure and specificity of a quorum-quenching lactonase (AiiB) from Agrobacterium tumefaciens.
Biochemistry, 46, 2007
|
|
4HCY
 
 | | Structure of a eukaryotic thiaminase-I bound to the thiamin analogue 3-deazathiamin | | 分子名称: | 2-{4-[(4-amino-2-methylpyrimidin-5-yl)methyl]-3-methylthiophen-2-yl}ethanol, thiaminase-I | | 著者 | Kreinbring, C.A, Hubbard, P.A, Leeper, F.J, Hawksley, D, Petsko, G.A, Ringe, D. | | 登録日 | 2012-10-01 | | 公開日 | 2013-10-02 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structure of a eukaryotic thiaminase I.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
