4R27
 
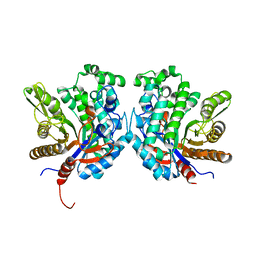 | | Crystal structure of beta-glycosidase BGL167 | | 分子名称: | Glycoside hydrolase | | 著者 | Park, S.J, Choi, J.M, Kyeong, H.H, Kim, S.G, Kim, H.S. | | 登録日 | 2014-08-09 | | 公開日 | 2015-05-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Rational design of a beta-glycosidase with high regiospecificity for triterpenoid tailoring
Chembiochem, 16, 2015
|
|
1YG0
 
 | |
1ZHC
 
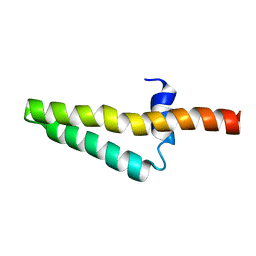 | |
1Z8M
 
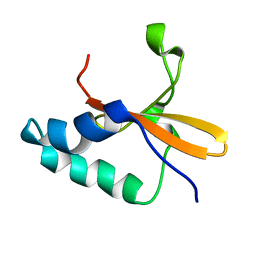 | |
3QJM
 
 | | Structural flexibility of Shank PDZ domain is important for its binding to different ligands | | 分子名称: | Beta-PIX, SH3 and multiple ankyrin repeat domains protein 1 | | 著者 | Lee, J.H, Park, H, Park, S.J, Kim, H.J, Eom, S.H. | | 登録日 | 2011-01-30 | | 公開日 | 2011-04-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.311 Å) | | 主引用文献 | The structural flexibility of the shank1 PDZ domain is important for its binding to different ligands
Biochem.Biophys.Res.Commun., 407, 2011
|
|
3QJN
 
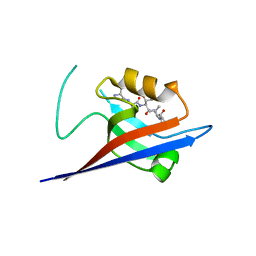 | | Structural flexibility of Shank PDZ domain is important for its binding to different ligands | | 分子名称: | Beta-PIX, SH3 and multiple ankyrin repeat domains protein 1 | | 著者 | Lee, J.H, Park, H, Park, S.J, Kim, H.J, Eom, S.H. | | 登録日 | 2011-01-30 | | 公開日 | 2011-04-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | The structural flexibility of the shank1 PDZ domain is important for its binding to different ligands
Biochem.Biophys.Res.Commun., 407, 2011
|
|
1Q3P
 
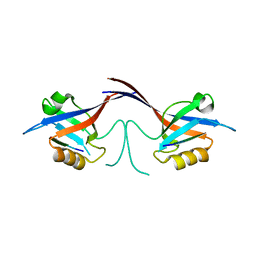 | | Crystal structure of the Shank PDZ-ligand complex reveals a class I PDZ interaction and a novel PDZ-PDZ dimerization | | 分子名称: | C-terminal hexapeptide from Guanylate kinase-associated protein, Shank1 | | 著者 | Im, Y.J, Lee, J.H, Park, S.H, Park, S.J, Rho, S.-H, Kang, G.B, Kim, E, Eom, S.H. | | 登録日 | 2003-07-31 | | 公開日 | 2004-01-27 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Crystal structure of the Shank PDZ-ligand complex reveals a class I PDZ interaction and a novel PDZ-PDZ dimerization
J.Biol.Chem., 278, 2003
|
|
1Q3O
 
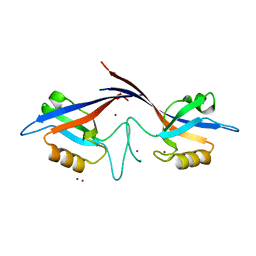 | | Crystal structure of the Shank PDZ-ligand complex reveals a class I PDZ interaction and a novel PDZ-PDZ dimerization | | 分子名称: | BROMIDE ION, Shank1 | | 著者 | Im, Y.J, Lee, J.H, Park, S.H, Park, S.J, Rho, S.-H, Kang, G.B, Kim, E, Eom, S.H. | | 登録日 | 2003-07-31 | | 公開日 | 2004-01-27 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of the Shank PDZ-ligand complex reveals a class I PDZ interaction and a novel PDZ-PDZ dimerization
J.Biol.Chem., 278, 2003
|
|
2OTR
 
 | |
2R62
 
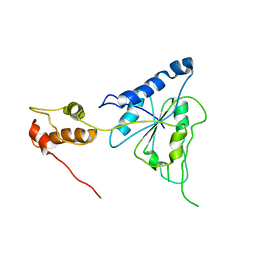 | | Crystal structure of Helicobacter pylori ATP dependent protease, FtsH | | 分子名称: | Cell division protease ftsH homolog | | 著者 | Kim, S.H, Kang, G.B, Song, H.-E, Park, S.J, Bae, M.-H, Eom, S.H. | | 登録日 | 2007-09-05 | | 公開日 | 2008-09-09 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structural studies on Helicobacter pyloriATP-dependent protease, FtsH
J.SYNCHROTRON RADIAT., 15, 2008
|
|
2R65
 
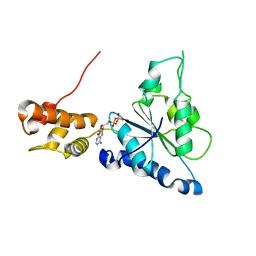 | | Crystal structure of Helicobacter pylori ATP dependent protease, FtsH ADP complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Cell division protease ftsH homolog | | 著者 | Kim, S.H, Kang, G.B, Song, H.-E, Park, S.J, Bae, M.-H, Eom, S.H. | | 登録日 | 2007-09-05 | | 公開日 | 2008-09-09 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structural studies on Helicobacter pyloriATP-dependent protease, FtsH
J.SYNCHROTRON RADIAT., 15, 2008
|
|
2H9Z
 
 | | Solution structure of hypothetical protein, HP0495 from Helicobacter pylori | | 分子名称: | Hypothetical protein HP0495 | | 著者 | Seo, M.D, Park, S.J, Kim, H.J, Lee, B.J. | | 登録日 | 2006-06-12 | | 公開日 | 2007-05-01 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of hypothetical protein, HP0495 (Y495_HELPY) from Helicobacter pylori.
Proteins, 67, 2007
|
|
9B8Y
 
 | |
9B93
 
 | |
9B92
 
 | |
9B91
 
 | |
9B8W
 
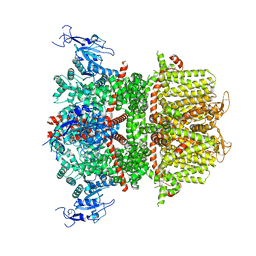 | |
9B90
 
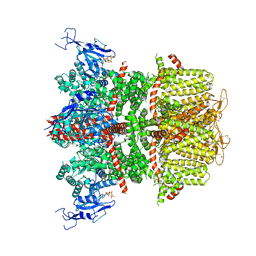 | |
9B8Z
 
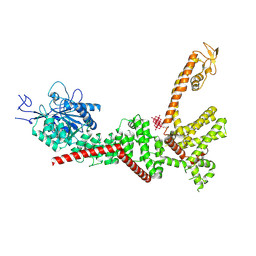 | |
9B8X
 
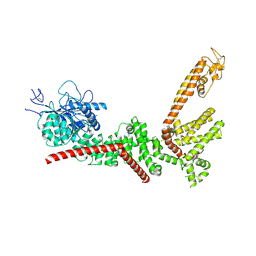 | |
9B94
 
 | |
6IFM
 
 | |
6IFC
 
 | | Crystal structure of VapBC from Salmonella typhimurium | | 分子名称: | Antitoxin VapB, CALCIUM ION, tRNA(fMet)-specific endonuclease VapC | | 著者 | Park, D.W, Lee, B.J. | | 登録日 | 2018-09-19 | | 公開日 | 2020-01-29 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Crystal structure of proteolyzed VapBC and DNA-bound VapBC from Salmonella enterica Typhimurium LT2 and VapC as a putative Ca2+-dependent ribonuclease.
Faseb J., 34, 2020
|
|
6IM5
 
 | | YAP-binding domain of human TEAD1 | | 分子名称: | PHOSPHATE ION, Transcriptional enhancer factor TEF-1 | | 著者 | Mo, Y, Lee, H.S, Kim, S.J, Ku, B. | | 登録日 | 2018-10-22 | | 公開日 | 2019-02-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.701 Å) | | 主引用文献 | Crystal Structure of the YAP-binding Domain of Human TEAD1
Bull.Korean Chem.Soc., 40, 2019
|
|
7CM4
 
 | | Crystal Structure of COVID-19 virus spike receptor-binding domain complexed with a neutralizing antibody CT-P59 | | 分子名称: | 1,2-ETHANEDIOL, IgG heavy chain, IgG light chain, ... | | 著者 | Kim, Y.G, Jeong, J.H, Bae, J.S, Lee, J. | | 登録日 | 2020-07-24 | | 公開日 | 2021-01-20 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | A therapeutic neutralizing antibody targeting receptor binding domain of SARS-CoV-2 spike protein.
Nat Commun, 12, 2021
|
|
