5G3Y
 
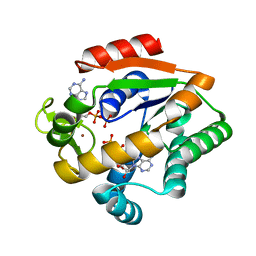 | | Crystal structure of adenylate kinase ancestor 1 with Zn and ADP bound | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENYLATE KINSE, ZINC ION | | 著者 | Nguyen, V, Kutter, S, English, J, Kern, D. | | 登録日 | 2016-05-03 | | 公開日 | 2016-12-28 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.18 Å) | | 主引用文献 | Evolutionary drivers of thermoadaptation in enzyme catalysis.
Science, 355, 2017
|
|
5G41
 
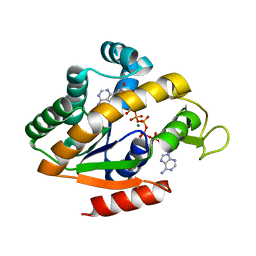 | | Crystal structure of adenylate kinase ancestor 4 with Zn, Mg and Ap5A bound | | 分子名称: | ADENYLATE KINSE, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Nguyen, V, Kutter, S, English, J, Kern, D. | | 登録日 | 2016-05-03 | | 公開日 | 2016-12-28 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Evolutionary drivers of thermoadaptation in enzyme catalysis.
Science, 355, 2017
|
|
5G40
 
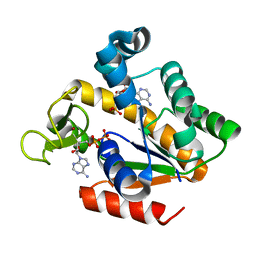 | | Crystal structure of adenylate kinase ancestor 4 with Zn and AMP-ADP bound | | 分子名称: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ADENYLATE KINSE, ... | | 著者 | Nguyen, V, Kutter, S, English, J, Kern, D. | | 登録日 | 2016-05-03 | | 公開日 | 2016-12-28 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Evolutionary drivers of thermoadaptation in enzyme catalysis.
Science, 355, 2017
|
|
5G3Z
 
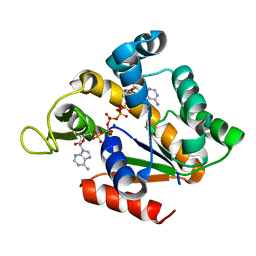 | | Crystal structure of adenylate kinase ancestor 3 with Zn, Mg and Ap5A bound | | 分子名称: | ADENYLATE KINSE, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Nguyen, V, Kutter, S, English, J, Kern, D. | | 登録日 | 2016-05-03 | | 公開日 | 2016-12-28 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Evolutionary drivers of thermoadaptation in enzyme catalysis.
Science, 355, 2017
|
|
8T7Q
 
 | | Identification of GDC-1971 (RLY-1971), a SHP2 inhibitor designed for the treatment of solid tumors | | 分子名称: | 1-{3-[(2-chlorophenyl)sulfanyl]-1H-pyrazolo[3,4-b]pyrazin-6-yl}-4-methylpiperidin-4-amine, Tyrosine-protein phosphatase non-receptor type 11 | | 著者 | Tang, Y, Nguyen, V, Wilbur, J.D. | | 登録日 | 2023-06-21 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Identification of GDC-1971 (RLY-1971), a SHP2 Inhibitor Designed for the Treatment of Solid Tumors.
J.Med.Chem., 66, 2023
|
|
8T8Q
 
 | | Identification of GDC-1971 (RLY-1971), a SHP2 inhibitor designed for the treatment of solid tumors | | 分子名称: | 1-[(3P)-3-(3-chloro-2-fluorophenyl)-1H-pyrazolo[3,4-b]pyrazin-6-yl]-4-methylpiperidin-4-amine, Tyrosine-protein phosphatase non-receptor type 11 | | 著者 | Tang, Y, Nguyen, V, Wilbur, J.D. | | 登録日 | 2023-06-23 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | Identification of GDC-1971 (RLY-1971), a SHP2 Inhibitor Designed for the Treatment of Solid Tumors.
J.Med.Chem., 66, 2023
|
|
8T6D
 
 | | Identification of GDC-1971 (RLY-1971), a SHP2 inhibitor designed for the treatment of solid tumors | | 分子名称: | (3R)-1'-[3-(3,4-dihydro-1,5-naphthyridin-1(2H)-yl)-1H-pyrazolo[3,4-b]pyrazin-6-yl]-3H-spiro[[1]benzofuran-2,4'-piperidin]-3-amine, SULFATE ION, Tyrosine-protein phosphatase non-receptor type 11 | | 著者 | Tang, Y, Nguyen, V, Wilbur, J.D. | | 登録日 | 2023-06-15 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Identification of GDC-1971 (RLY-1971), a SHP2 Inhibitor Designed for the Treatment of Solid Tumors.
J.Med.Chem., 66, 2023
|
|
8G4L
 
 | | Cryo-EM structure of the human cardiac myosin filament | | 分子名称: | Myosin light chain 3, Myosin regulatory light chain 2, ventricular/cardiac muscle isoform, ... | | 著者 | Dutta, D, Nguyen, V, Padron, R, Craig, R. | | 登録日 | 2023-02-10 | | 公開日 | 2023-11-01 | | 最終更新日 | 2023-12-06 | | 実験手法 | ELECTRON MICROSCOPY (6.4 Å) | | 主引用文献 | Cryo-EM structure of the human cardiac myosin filament.
Nature, 623, 2023
|
|
6C83
 
 | | Structure of Aurora A (122-403) bound to inhibitory Monobody Mb2 and AMPPCP | | 分子名称: | Aurora kinase A, Mb2 Monobody, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | 著者 | Hoemberger, M, Kutter, S, Zorba, A, Nguyen, V, Shohei, A, Shohei, K, Kern, D. | | 登録日 | 2018-01-24 | | 公開日 | 2019-02-27 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Allosteric modulation of a human protein kinase with monobodies.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7K4R
 
 | | Crystal structure of Kemp Eliminase HG3 K50Q | | 分子名称: | Endo-1,4-beta-xylanase | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
7K4T
 
 | | Crystal structure of Kemp Eliminase HG3.17 | | 分子名称: | Endo-1,4-beta-xylanase | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (0.999 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
7K4P
 
 | | Crystal structure of Kemp Eliminase HG3 | | 分子名称: | Endo-1,4-beta-xylanase | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.08 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
7K4S
 
 | | Crystal structure of Kemp Eliminase HG3.7 | | 分子名称: | Endo-1,4-beta-xylanase, SULFATE ION | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
7K4X
 
 | | Crystal structure of Kemp Eliminase HG3.7 in complex with the transition state analog 6-nitrobenzotriazole | | 分子名称: | 6-NITROBENZOTRIAZOLE, ACETATE ION, Endo-1,4-beta-xylanase, ... | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
7K4Z
 
 | | Crystal structure of Kemp Eliminase HG3.17 in complex with the transition state analog 6-nitrobenzotriazole | | 分子名称: | 6-NITROBENZOTRIAZOLE, Endo-1,4-beta-xylanase, PENTAETHYLENE GLYCOL | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.08 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
7K4Q
 
 | | Crystal structure of Kemp Eliminase HG3 in complex with the transition state analog 6-nitrobenzotriazole | | 分子名称: | 6-NITROBENZOTRIAZOLE, Endo-1,4-beta-xylanase, SULFATE ION | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.28 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
7K4V
 
 | | Crystal structure of Kemp Eliminase HG3.17 | | 分子名称: | Endo-1,4-beta-xylanase, TETRAETHYLENE GLYCOL | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
7K4W
 
 | | Crystal structure of Kemp Eliminase HG3.17 in the inactive state | | 分子名称: | CALCIUM ION, Endo-1,4-beta-xylanase | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
7K4U
 
 | | Crystal structure of Kemp Eliminase HG3 K50Q in complex with the transition state analog 6-nitrobenzotriazole | | 分子名称: | 6-NITROBENZOTRIAZOLE, Endo-1,4-beta-xylanase | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
7K4Y
 
 | | Crystal structure of Kemp Eliminase HG3.17 at 343 K | | 分子名称: | Endo-1,4-beta-xylanase | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
4GMR
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR266. | | 分子名称: | NITRATE ION, OR266 DE NOVO PROTEIN | | 著者 | Vorobiev, S, Su, M, Parmeggiani, F, Seetharaman, J, Huang, P.-S, Mao, M, Xiao, R, Lee, D, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2012-08-16 | | 公開日 | 2012-08-29 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.377 Å) | | 主引用文献 | Computational design of self-assembling cyclic protein homo-oligomers.
NAT.CHEM., 9, 2017
|
|
4GPM
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR264. | | 分子名称: | Engineered Protein OR264 | | 著者 | Vorobiev, S, Su, M, Parmeggiani, F, Seetharaman, J, Huang, P.-S, Maglaqui, M, Xiao, R, Lee, D, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2012-08-21 | | 公開日 | 2012-09-05 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.002 Å) | | 主引用文献 | Computational design of self-assembling cyclic protein homo-oligomers.
NAT.CHEM., 9, 2017
|
|
5HS0
 
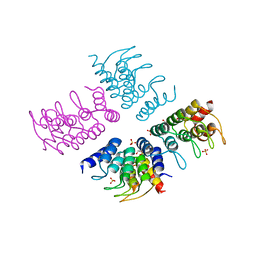 | | Computationally Designed Cyclic Tetramer ank1C4_7 | | 分子名称: | Ankyrin domain protein ank1C4_7, GLYCEROL, SULFATE ION | | 著者 | McNamara, D.E, Cascio, D, Fallas, J.A, Baker, D, Yeates, T.O. | | 登録日 | 2016-01-24 | | 公開日 | 2017-04-12 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Computational design of self-assembling cyclic protein homo-oligomers.
Nat Chem, 9, 2017
|
|
5HRZ
 
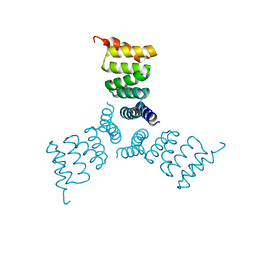 | | Computationally Designed Trimer 1na0C3_3 | | 分子名称: | TPR domain protein 1na0C3_3 | | 著者 | Cascio, D, McNamara, D.E, Fallas, J.A, Baker, D, Yeates, T.O. | | 登録日 | 2016-01-24 | | 公開日 | 2017-04-12 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Computational design of self-assembling cyclic protein homo-oligomers.
Nat Chem, 9, 2017
|
|
8T6G
 
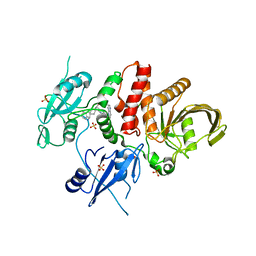 | | Identification of GDC-1971 (RLY-1971), a SHP2 inhibitor designed for the treatment of solid tumors | | 分子名称: | (1S)-1-{6-[(1S)-1-amino-1,3-dihydrospiro[indene-2,4'-piperidin]-1'-yl]-3-(3,4-dihydro-1,5-naphthyridin-1(2H)-yl)-1H-pyrazolo[3,4-b]pyrazin-5-yl}ethan-1-ol, SULFATE ION, Tyrosine-protein phosphatase non-receptor type 11 | | 著者 | Tang, Y, Nugyen, V, Wilbur, J.D. | | 登録日 | 2023-06-15 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Identification of GDC-1971 (RLY-1971), a SHP2 Inhibitor Designed for the Treatment of Solid Tumors.
J.Med.Chem., 66, 2023
|
|
