4PDW
 
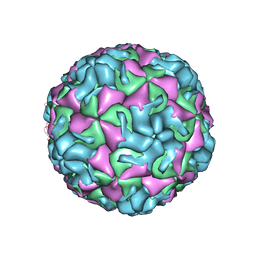 | | A benzonitrile analogue inhibits rhinovirus replication | | 分子名称: | 4-[(4,5-dimethoxy-2-nitrophenyl)acetyl]benzonitrile, Capsid protein VP4/VP2, GLYCEROL, ... | | 著者 | Querol-Audi, J, Guerra, P, Lacroix, C, Roche, M, Franco, D, Froeyen, M, Terme, T, Vanelle, P, Neyts, J, Leyssen, P, Verdaguer, N. | | 登録日 | 2014-04-22 | | 公開日 | 2014-07-02 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | A novel benzonitrile analogue inhibits rhinovirus replication.
J.Antimicrob.Chemother., 69, 2014
|
|
6DIZ
 
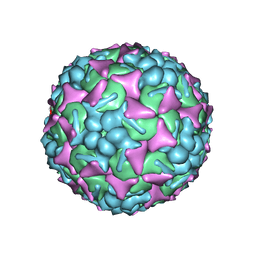 | | EV-A71 strain 11316 complexed with tryptophan dendrimer MADAL_0385 | | 分子名称: | SPHINGOSINE, VP1, VP2, ... | | 著者 | Sun, L, Lee, H, Thibaut, H.J, Rivero-Buceta, E, Martinez-Gualda, B, Delang, L, Leyssen, P, Gago, F, San-Felix, A, Hafenstein, S, Mirabelli, C, Neyts, J. | | 登録日 | 2018-05-24 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.59 Å) | | 主引用文献 | Viral engagement with host (co-)receptors blocked by a novel class of tryptophan dendrimers that targets the 5-fold-axis of the enterovirus-A71 capsid.
Plos Pathog., 15, 2019
|
|
5ABJ
 
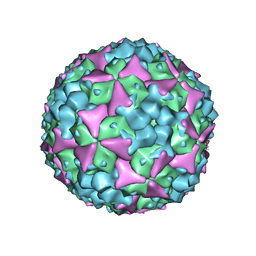 | | Structure of Coxsackievirus A16 in complex with GPP3 | | 分子名称: | 1-[(3S)-5-[4-[(E)-ETHOXYIMINOMETHYL]PHENOXY]-3-METHYL-PENTYL]-3-PYRIDIN-4-YL-IMIDAZOLIDIN-2-ONE, CHLORIDE ION, SODIUM ION, ... | | 著者 | De Colibus, L, Wang, X, Tijsma, A, Neyts, J, Spyrou, J.A.B, Ren, J, Grimes, J.M, Puerstinger, G, Leyssen, P, Fry, E.E, Rao, Z, Stuart, D.I. | | 登録日 | 2015-08-06 | | 公開日 | 2015-09-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structure Elucidation of Coxsackievirus A16 in Complex with Gpp3 Informs a Systematic Review of Highly Potent Capsid Binders to Enteroviruses.
Plos Pathog., 11, 2015
|
|
6FV1
 
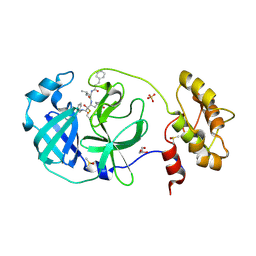 | | Structure of human coronavirus NL63 main protease in complex with the alpha-ketoamide (S)-N-((S)-4-(benzylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)butan-2-yl)-2-cinnamamido-4-methylpentanamide (cinnamoyl-leucine-GlnLactam-CO-CO-NH-benzyl) | | 分子名称: | (2~{S})-4-methyl-~{N}-[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]-2-[[(~{E})-3-phenylprop-2-enoyl]amino]pentanamide, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | 著者 | Zhang, L, Hilgenfeld, R. | | 登録日 | 2018-02-28 | | 公開日 | 2019-03-20 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Alpha-ketoamides as broad-spectrum inhibitors of coronavirus and enterovirus replication Structure-based design, synthesis, and activity assessment.
J.Med.Chem., 2020
|
|
7B3E
 
 | | Crystal structure of myricetin covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 | | 分子名称: | 1,2-ETHANEDIOL, 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, CHLORIDE ION, ... | | 著者 | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | 登録日 | 2020-11-30 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Identification of Inhibitors of SARS-CoV-2 3CL-Pro Enzymatic Activity Using a Small Molecule in Vitro Repurposing Screen.
Acs Pharmacol Transl Sci, 4, 2021
|
|
7SL5
 
 | |
7SKZ
 
 | |
8ERR
 
 | |
8ERQ
 
 | |
7TAS
 
 | |
7TAT
 
 | |
7O46
 
 | |
7P40
 
 | | P5C3 is a potent fab neutralizer | | 分子名称: | Spike glycoprotein, Variable Heavy Chain P5C3 (VH), Variable Light Chain P5C3 (VL) | | 著者 | perez, L. | | 登録日 | 2021-07-09 | | 公開日 | 2021-10-13 | | 最終更新日 | 2023-04-12 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | A highly potent antibody effective against SARS-CoV-2 variants of concern.
Cell Rep, 37, 2021
|
|
7PHG
 
 | | MaP OF P5C3RBD Interface | | 分子名称: | Heavy ChaIn variable, Light ChaIn, Surface glycoprotein | | 著者 | Perez, L. | | 登録日 | 2021-08-17 | | 公開日 | 2021-10-13 | | 最終更新日 | 2021-10-27 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | A highly potent antibody effective against SARS-CoV-2 variants of concern.
Cell Rep, 37, 2021
|
|
7U0A
 
 | |
7U09
 
 | |
7U0E
 
 | |
8A4T
 
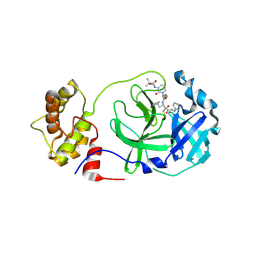 | | crystal structures of diastereomer (S,S,S)-13b (13b-K) in complex with the SARS-CoV-2 Mpro | | 分子名称: | 3C-like proteinase nsp5, CHLORIDE ION, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | 著者 | Ibrahim, M, Hilgenfeld, R, Zhang, L. | | 登録日 | 2022-06-13 | | 公開日 | 2022-10-12 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Diastereomeric Resolution Yields Highly Potent Inhibitor of SARS-CoV-2 Main Protease.
J.Med.Chem., 65, 2022
|
|
8A4Q
 
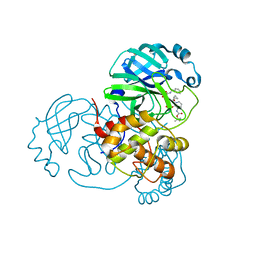 | | crystal structures of diastereomer (R,S,S)-13b (13b-H) in complex with the SARS-CoV-2 Mpro. | | 分子名称: | 3C-like proteinase nsp5, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Ibrahim, M, Hilgenfeld, R, Zhang, L. | | 登録日 | 2022-06-13 | | 公開日 | 2022-10-12 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Diastereomeric Resolution Yields Highly Potent Inhibitor of SARS-CoV-2 Main Protease.
J.Med.Chem., 65, 2022
|
|
6FV2
 
 | |
8QPR
 
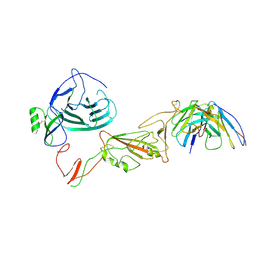 | | SARS-CoV-2 S protein bound to human neutralising antibody UZGENT_G5 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, IgG heavy chain - FAB, ... | | 著者 | Remaut, H, Reiter, D, Vandenkerckhove, L, Acar, D.D, Witkowski, W, Gerlo, S. | | 登録日 | 2023-10-03 | | 公開日 | 2024-01-31 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Integrating artificial intelligence-based epitope prediction in a SARS-CoV-2 antibody discovery pipeline: caution is warranted.
Ebiomedicine, 100, 2024
|
|
8QQ0
 
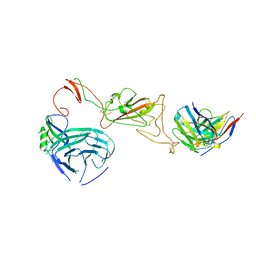 | | SARS-CoV-2 S protein bound to neutralising antibody UZGENT_A3 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Remaut, H, Reiter, D, Vandenkerckhove, L, Acar, D.D, Witkowski, W, Gerlo, S. | | 登録日 | 2023-10-03 | | 公開日 | 2024-01-31 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Integrating artificial intelligence-based epitope prediction in a SARS-CoV-2 antibody discovery pipeline: caution is warranted.
Ebiomedicine, 100, 2024
|
|
3CDU
 
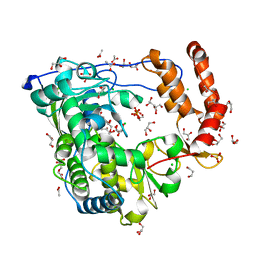 | | Crystal structure of coxsackievirus B3 RNA-dependent RNA polymerase (3Dpol) in complex with a pyrophosphate | | 分子名称: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | 著者 | Gruez, A, Selisko, B, Roberts, M, Bricogne, G, Bussetta, C, Canard, B. | | 登録日 | 2008-02-27 | | 公開日 | 2008-07-22 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The crystal structure of coxsackievirus B3 RNA-dependent RNA polymerase in complex with its protein primer VPg confirms the existence of a second VPg binding site on Picornaviridae polymerases
J.Virol., 82, 2008
|
|
3CDW
 
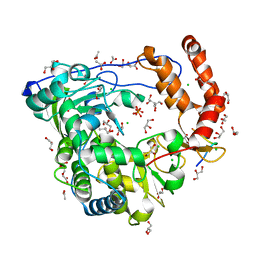 | | Crystal structure of coxsackievirus B3 RNA-dependent RNA polymerase (3Dpol) in complex with protein primer VPg and a pyrophosphate | | 分子名称: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | 著者 | Gruez, A, Selisko, B, Roberts, M, Bricogne, G, Bussetta, C, Canard, B. | | 登録日 | 2008-02-27 | | 公開日 | 2008-07-22 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The crystal structure of coxsackievirus B3 RNA-dependent RNA polymerase in complex with its protein primer VPg confirms the existence of a second VPg binding site on Picornaviridae polymerases
J.Virol., 82, 2008
|
|
6GZV
 
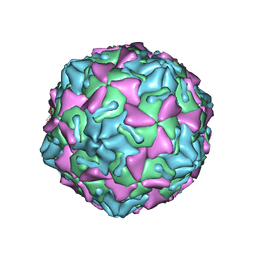 | | Identification of a druggable VP1-VP3 interprotomer pocket in the capsid of enteroviruses | | 分子名称: | 4-[[4-[1,3-bis(oxidanylidene)isoindol-2-yl]phenyl]sulfonylamino]benzoic acid, Capsid protein VP1, Capsid protein VP2, ... | | 著者 | Geraets, J.A, Flatt, J.W, Domanska, A, Butcher, S.J. | | 登録日 | 2018-07-05 | | 公開日 | 2019-06-05 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | A novel druggable interprotomer pocket in the capsid of rhino- and enteroviruses.
Plos Biol., 17, 2019
|
|
