6AK3
 
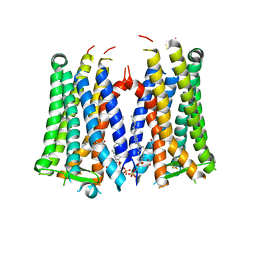 | | Crystal structure of the human prostaglandin E receptor EP3 bound to prostaglandin E2 | | 分子名称: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxo-cyclopentyl]hept-5-enoic acid, Prostaglandin E2 receptor EP3 subtype,Soluble cytochrome b562 | | 著者 | Morimoto, K, Suno, R, Iwata, S, Kobayashi, T. | | 登録日 | 2018-08-29 | | 公開日 | 2018-12-05 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structure of the endogenous agonist-bound prostanoid receptor EP3.
Nat. Chem. Biol., 15, 2019
|
|
1X0G
 
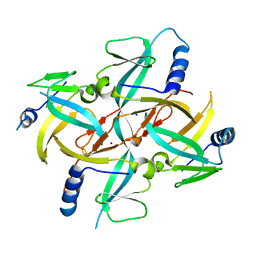 | | Crystal Structure of IscA with the [2Fe-2S] cluster | | 分子名称: | FE2/S2 (INORGANIC) CLUSTER, IscA, SODIUM ION | | 著者 | Morimoto, K, Yamashita, E, Kondou, Y, Lee, S.J, Tsukihara, T, Nakai, M. | | 登録日 | 2005-03-22 | | 公開日 | 2006-06-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The Asymmetric IscA Homodimer with an Exposed [2Fe-2S] Cluster Suggests the Structural Basis of the Fe-S Cluster Biosynthetic Scaffold.
J.Mol.Biol., 360, 2006
|
|
6JT4
 
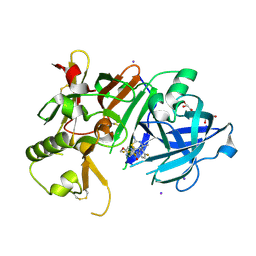 | | Crystal Structure of BACE1 in complex with N-{3-[(4S,6S)-2-amino-4-methyl-6-(trifluoromethyl)-5,6-dihydro-4H-1,3-thiazin-4-yl]-4-fluorophenyl}-5-(fluoromethoxy)pyrazine-2-carboxamide | | 分子名称: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | 著者 | Anan, K, Iso, Y, Oguma, T, Nakahara, K, Suzuki, S, Yamamoto, T, Matsuoka, E, Ito, H, Sakaguchi, G, Ando, S, Morimoto, K, Kanegawa, N, Kido, Y, Kawachi, T, Fukushima, T, Teisman, A, Urmaliya, V, Dhuyvetter, D, Borghys, H, Austin, N, Bergh, A.V.D, Verboven, P, Bischoff, F, Gijsen, H.J.M, Yamano, Y, Kusakabe, K.I. | | 登録日 | 2019-04-08 | | 公開日 | 2020-01-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Trifluoromethyl Dihydrothiazine-Based beta-Secretase (BACE1) Inhibitors with Robust Central beta-Amyloid Reduction and Minimal Covalent Binding Burden.
Chemmedchem, 14, 2019
|
|
7WU9
 
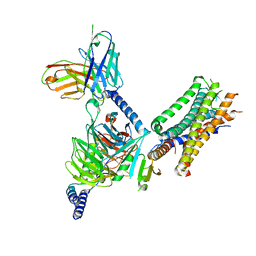 | | Cryo-EM structure of the human EP3-Gi signaling complex | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | 著者 | Suno, R, Sugita, Y, Morimoto, K, Iwasaki, K, Kato, T, Kobayashi, T. | | 登録日 | 2022-02-07 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-10-04 | | 実験手法 | ELECTRON MICROSCOPY (3.375 Å) | | 主引用文献 | Structural insights into the G protein selectivity revealed by the human EP3-G i signaling complex.
Cell Rep, 40, 2022
|
|
7DCZ
 
 | | Crystal Structure of BACE1 in complex with N-{3-[(4S)-2-amino-4-methyl-4H-1,3-thiazin-4-yl]-4- fluorophenyl}-5-cyanopyridine-2-carboxamide | | 分子名称: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | 著者 | Koriyama, Y, Hori, A, Ito, H, Yonezawa, S, Baba, Y, Tanimoto, N, Ueno, T, Yamamoto, S, Yamamoto, T, Asada, N, Morimoto, K, Einaru, S, Sakai, K, Kanazu, T, Matsuda, A, Yamaguchi, Y, Oguma, T, Timmers, M, Tritsmans, L, Kusakabe, K.I, Kato, A, Sakaguchi, G. | | 登録日 | 2020-10-27 | | 公開日 | 2021-03-10 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Discovery of Atabecestat (JNJ-54861911): A Thiazine-Based beta-Amyloid Precursor Protein Cleaving Enzyme 1 Inhibitor Advanced to the Phase 2b/3 EARLY Clinical Trial.
J.Med.Chem., 64, 2021
|
|
5YFI
 
 | | Crystal structure of the anti-human prostaglandin E receptor EP4 antibody Fab fragment | | 分子名称: | Heavy chain of Fab fragment, Light chain of Fab fragment, ZINC ION | | 著者 | Toyoda, Y, Morimoto, K, Suno, R, Horita, S, Iwata, S, Kobayashi, T. | | 登録日 | 2017-09-21 | | 公開日 | 2018-12-05 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.848 Å) | | 主引用文献 | Ligand binding to human prostaglandin E receptor EP4at the lipid-bilayer interface.
Nat. Chem. Biol., 15, 2019
|
|
5YHL
 
 | | Crystal structure of the human prostaglandin E receptor EP4 in complex with Fab and an antagonist Br-derivative | | 分子名称: | 4-[2-[[(2R)-2-(4-bromanylnaphthalen-1-yl)propanoyl]amino]-4-cyano-phenyl]butanoic acid, Heavy chain of Fab fragment, Light chain of Fab fragment, ... | | 著者 | Toyoda, Y, Morimoto, K, Suno, R, Horita, S, Iwata, S, Kobayashi, T. | | 登録日 | 2017-09-28 | | 公開日 | 2018-12-05 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (4.2 Å) | | 主引用文献 | Ligand binding to human prostaglandin E receptor EP4at the lipid-bilayer interface.
Nat. Chem. Biol., 15, 2019
|
|
5YWY
 
 | | Crystal structure of the human prostaglandin E receptor EP4 in complex with Fab and ONO-AE3-208 | | 分子名称: | 4-[4-cyano-2-[[(2R)-2-(4-fluoranylnaphthalen-1-yl)propanoyl]amino]phenyl]butanoic acid, Heavy chain of Fab fragment, Light chain of Fab fragment, ... | | 著者 | Toyoda, Y, Morimoto, K, Suno, R, Horita, S, Iwata, S, Kobayashi, T. | | 登録日 | 2017-11-30 | | 公開日 | 2018-12-05 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Ligand binding to human prostaglandin E receptor EP4at the lipid-bilayer interface.
Nat. Chem. Biol., 15, 2019
|
|
7D7M
 
 | | Cryo-EM Structure of the Prostaglandin E Receptor EP4 Coupled to G Protein | | 分子名称: | (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxo-cyclopentyl]hept-5-enoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Nojima, S, Fujita, Y, Kimura, T.K, Nomura, N, Suno, R, Morimoto, K, Yamamoto, M, Noda, T, Iwata, S, Shigematsu, H, Kobayashi, T. | | 登録日 | 2020-10-05 | | 公開日 | 2020-11-18 | | 最終更新日 | 2021-03-17 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Cryo-EM Structure of the Prostaglandin E Receptor EP4 Coupled to G Protein.
Structure, 29, 2021
|
|
1FLM
 
 | | DIMER OF FMN-BINDING PROTEIN FROM DESULFOVIBRIO VULGARIS (MIYAZAKI F) | | 分子名称: | FLAVIN MONONUCLEOTIDE, PROTEIN (FMN-BINDING PROTEIN) | | 著者 | Suto, K, Kawagoe, K, Shibata, N, Morimoto, K, Higuchi, Y, Kitamura, M, Nakaya, T, Yasuoka, N. | | 登録日 | 1999-03-10 | | 公開日 | 2000-03-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | How do the x-ray structure and the NMR structure of FMN-binding protein differ?
Acta Crystallogr.,Sect.D, 56, 2000
|
|
7N4N
 
 | | BACE-2 in complex with ligand 36 | | 分子名称: | 1,2-ETHANEDIOL, Beta-secretase 2, N-{3-[(2S,5R)-6-amino-2-(fluoromethyl)-5-(methanesulfonyl)-5-methyl-2,3,4,5-tetrahydropyridin-2-yl]-4-fluorophenyl}-6-methoxypyrimidine-4-carboxamide, ... | | 著者 | Shaffer, P.L. | | 登録日 | 2021-06-04 | | 公開日 | 2021-10-06 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (1.41 Å) | | 主引用文献 | JNJ-67569762, A 2-Aminotetrahydropyridine-Based Selective BACE1 Inhibitor Targeting the S3 Pocket: From Discovery to Clinical Candidate.
J.Med.Chem., 64, 2021
|
|
7N66
 
 | | BACE-1 in complex with ligand 12 | | 分子名称: | Beta-secretase 1, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Shaffer, P.L. | | 登録日 | 2021-06-07 | | 公開日 | 2021-10-06 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | JNJ-67569762, A 2-Aminotetrahydropyridine-Based Selective BACE1 Inhibitor Targeting the S3 Pocket: From Discovery to Clinical Candidate.
J.Med.Chem., 64, 2021
|
|
7DC0
 
 | |
7DC4
 
 | | Crystal structure of glycan-bound Pseudomonas taiwanensis lectin | | 分子名称: | Lectin, SULFATE ION, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose | | 著者 | Oda, K, Matoba, Y. | | 登録日 | 2020-10-23 | | 公開日 | 2021-04-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (0.95 Å) | | 主引用文献 | Lectins engineered to favor a glycan-binding conformation have enhanced antiviral activity.
J.Biol.Chem., 296, 2021
|
|
3VYL
 
 | | Structure of L-ribulose 3-epimerase | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, L-ribulose 3-epimerase, MANGANESE (II) ION | | 著者 | Uechi, K, Sakuraba, H, Takata, G. | | 登録日 | 2012-09-27 | | 公開日 | 2013-10-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural insight into L-ribulose 3-epimerase from Mesorhizobium loti.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3WW3
 
 | | X-ray structures of Cellulomonas parahominis L-ribose isomerase with no ligand | | 分子名称: | L-ribose isomerase, MANGANESE (II) ION | | 著者 | Terami, Y, Yoshida, H, Takata, G, Kamitori, S. | | 登録日 | 2014-06-13 | | 公開日 | 2015-04-29 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Essentiality of tetramer formation of Cellulomonas parahominis L-ribose isomerase involved in novel L-ribose metabolic pathway.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
3WW1
 
 | | X-ray structure of Cellulomonas parahominis L-ribose isomerase with L-ribose | | 分子名称: | L-ribose, L-ribose isomerase, MANGANESE (II) ION, ... | | 著者 | Terami, Y, Yoshida, H, Takata, G, Kamitori, S. | | 登録日 | 2014-06-13 | | 公開日 | 2015-04-29 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Essentiality of tetramer formation of Cellulomonas parahominis L-ribose isomerase involved in novel L-ribose metabolic pathway.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
3WW2
 
 | | X-ray structures of Cellulomonas parahominis L-ribose isomerase with L-psicose | | 分子名称: | L-psicose, L-ribose isomerase, MANGANESE (II) ION, ... | | 著者 | Terami, Y, Yoshida, H, Takata, G, Kamitori, S. | | 登録日 | 2014-06-13 | | 公開日 | 2015-04-29 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Essentiality of tetramer formation of Cellulomonas parahominis L-ribose isomerase involved in novel L-ribose metabolic pathway.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
3WW4
 
 | | X-ray structures of Cellulomonas parahominis L-ribose isomerase with L-allose | | 分子名称: | L-allose, L-ribose isomerase, MANGANESE (II) ION, ... | | 著者 | Terami, Y, Yoshida, H, Takata, G, Kamitori, S. | | 登録日 | 2014-06-13 | | 公開日 | 2015-04-29 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Essentiality of tetramer formation of Cellulomonas parahominis L-ribose isomerase involved in novel L-ribose metabolic pathway.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
2Z51
 
 | |
