4RYU
 
 | |
4RYT
 
 | |
4G9O
 
 | |
4GAD
 
 | |
6XEU
 
 | | CryoEM structure of GIRK2PIP2* - G protein-gated inwardly rectifying potassium channel GIRK2 with PIP2 | | 分子名称: | G protein-activated inward rectifier potassium channel 2, POTASSIUM ION, SODIUM ION, ... | | 著者 | Mathiharan, Y.K, Glaaser, I.W, Skiniotis, G, Slesinger, P.A. | | 登録日 | 2020-06-13 | | 公開日 | 2021-09-01 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural insights into GIRK2 channel modulation by cholesterol and PIP2
Cell Rep, 36, 2021
|
|
6XEV
 
 | | CryoEM structure of GIRK2-PIP2/CHS - G protein-gated inwardly rectifying potassium channel GIRK2 with modulators cholesteryl hemisuccinate and PIP2 | | 分子名称: | CHOLESTEROL HEMISUCCINATE, G protein-activated inward rectifier potassium channel 2, POTASSIUM ION, ... | | 著者 | Mathiharan, Y.K, Glaaser, I.W, Skiniotis, G, Slesinger, P.A. | | 登録日 | 2020-06-14 | | 公開日 | 2021-09-01 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural insights into GIRK2 channel modulation by cholesterol and PIP2
Cell Rep, 36, 2021
|
|
4XGB
 
 | | Crystal Structure of E112A/H234A Mutant of Stationary Phase Survival Protein (SurE) from Salmonella typhimurium co-crystallized with AMP | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5'/3'-nucleotidase SurE, MAGNESIUM ION, ... | | 著者 | Mathiharan, Y.K, Murthy, M.R.N. | | 登録日 | 2014-12-30 | | 公開日 | 2015-09-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | Insights into stabilizing interactions in the distorted domain-swapped dimer of Salmonella typhimurium survival protein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XGP
 
 | | Crystal Structure of E112A/H234A Mutant of Stationary Phase Survival Protein (SurE) from Salmonella typhimurium co-crystallized and soaked with AMP. | | 分子名称: | 1,2-ETHANEDIOL, 5'/3'-nucleotidase SurE, ADENINE, ... | | 著者 | Mathiharan, Y.K, Murthy, M.R.N. | | 登録日 | 2015-01-01 | | 公開日 | 2015-09-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Insights into stabilizing interactions in the distorted domain-swapped dimer of Salmonella typhimurium survival protein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XEP
 
 | | Crystal Structure of F222 form of E112A/H234A Mutant of Stationary Phase Survival Protein (SurE) from Salmonella typhimurium | | 分子名称: | 1,2-ETHANEDIOL, 5'/3'-nucleotidase SurE, MAGNESIUM ION, ... | | 著者 | Mathiharan, Y.K, Murthy, M.R.N. | | 登録日 | 2014-12-24 | | 公開日 | 2015-09-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Insights into stabilizing interactions in the distorted domain-swapped dimer of Salmonella typhimurium survival protein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XJ7
 
 | |
4XER
 
 | | Crystal Structure of C2 form of E112A/H234A Mutant of Stationary Phase Survival Protein (SurE) from Salmonella typhimurium | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5'/3'-nucleotidase SurE, ACETATE ION, ... | | 著者 | Mathiharan, Y.K, Murthy, M.R.N. | | 登録日 | 2014-12-24 | | 公開日 | 2015-09-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Insights into stabilizing interactions in the distorted domain-swapped dimer of Salmonella typhimurium survival protein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XH8
 
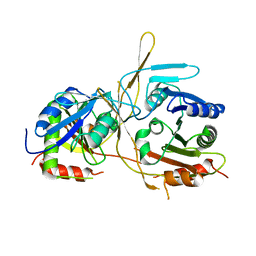 | |
8T6V
 
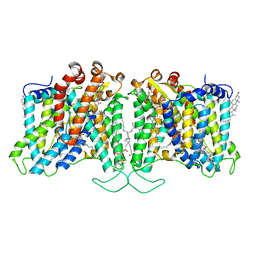 | | Cryo-EM structure of human Anion Exchanger 1 bound to 4,4'-Diisothiocyanatostilbene-2,2'-Disulfonic Acid (DIDS) | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4,4'-Diisothiocyano-2,2'-stilbenedisulfonic acid, ... | | 著者 | Capper, M.J, Zilberg, G, Mathiharan, Y.K, Yang, S, Stone, A.C, Wacker, D. | | 登録日 | 2023-06-18 | | 公開日 | 2023-09-13 | | 最終更新日 | 2023-11-01 | | 実験手法 | ELECTRON MICROSCOPY (2.95 Å) | | 主引用文献 | Substrate binding and inhibition of the anion exchanger 1 transporter.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8T6U
 
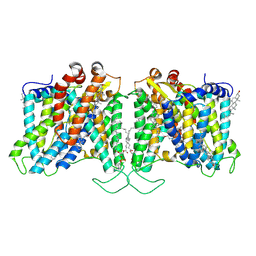 | | Cryo-EM structure of human Anion Exchanger 1 bound to Dipyridamole | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-[[2-[bis(2-hydroxyethyl)amino]-4,8-di(piperidin-1-yl)pyrimido[5,4-d]pyrimidin-6-yl]-(2-hydroxyethyl)amino]ethanol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Capper, M.J, Zilberg, G, Mathiharan, Y.K, Yang, S, Stone, A.C, Wacker, D. | | 登録日 | 2023-06-18 | | 公開日 | 2023-09-13 | | 最終更新日 | 2023-11-01 | | 実験手法 | ELECTRON MICROSCOPY (3.13 Å) | | 主引用文献 | Substrate binding and inhibition of the anion exchanger 1 transporter.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5XN8
 
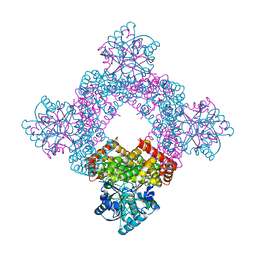 | | Structure of glycerol dehydrogenase crystallised as a contaminant | | 分子名称: | GLYCEROL, Glycerol Dehydrogenase, ZINC ION | | 著者 | Hatti, K, Mathiharan, Y.K, Srinivasan, N, Murthy, M.R.N. | | 登録日 | 2017-05-19 | | 公開日 | 2017-06-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | Seeing but not believing: the structure of glycerol dehydrogenase initially assumed to be the structure of a survival protein from Salmonella typhimurium
Acta Crystallogr.,Sect.D, 73, 2017
|
|
6B19
 
 | | Architecture of HIV-1 reverse transcriptase initiation complex core | | 分子名称: | RNA genome fragment, reverse transcriptase p51 subunit, reverse transcriptase p66 subunit, ... | | 著者 | Larsen, K.P, Mathiharan, Y.K, Chen, D.H, Puglisi, J.D, Skiniotis, G, Puglisi, E.V. | | 登録日 | 2017-09-18 | | 公開日 | 2018-04-25 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Architecture of an HIV-1 reverse transcriptase initiation complex.
Nature, 557, 2018
|
|
7TY7
 
 | | Cryo-EM structure of human Anion Exchanger 1 bound to Bicarbonate | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BICARBONATE ION, ... | | 著者 | Capper, M.J, Mathiharan, Y.K, Yang, S, Stone, A.C, Wacker, D. | | 登録日 | 2022-02-11 | | 公開日 | 2023-08-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | ELECTRON MICROSCOPY (3.37 Å) | | 主引用文献 | Substrate binding and inhibition of the anion exchanger 1 transporter.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7TY6
 
 | | Cryo-EM structure of human Anion Exchanger 1 bound to 4,4'-Diisothiocyanatodihydrostilbene-2,2'-Disulfonic Acid (H2DIDS) | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4,4'-Diisothiocyano-2,2'-stilbenedisulfonic acid, ... | | 著者 | Capper, M.J, Mathiharan, Y.K, Yang, S, Stone, A.C, Wacker, D. | | 登録日 | 2022-02-11 | | 公開日 | 2023-08-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | ELECTRON MICROSCOPY (2.98 Å) | | 主引用文献 | Substrate binding and inhibition of the anion exchanger 1 transporter.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7TY4
 
 | | Cryo-EM structure of human Anion Exchanger 1 | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Band 3 anion transport protein, ... | | 著者 | Capper, M.J, Mathiharan, Y.K, Yang, S, Stone, A.C, Wacker, D. | | 登録日 | 2022-02-11 | | 公開日 | 2023-08-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | ELECTRON MICROSCOPY (2.99 Å) | | 主引用文献 | Substrate binding and inhibition of the anion exchanger 1 transporter.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7TYA
 
 | | Cryo-EM structure of human Anion Exchanger 1 modified with Diethyl Pyrocarbonate (DEPC) | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Band 3 anion transport protein, ... | | 著者 | Capper, M.J, Mathiharan, Y.K, Yang, S, Stone, A.C, Wacker, D. | | 登録日 | 2022-02-11 | | 公開日 | 2023-08-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | Substrate binding and inhibition of the anion exchanger 1 transporter.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7TY8
 
 | | Cryo-EM structure of human Anion Exchanger 1 bound to Niflumic Acid | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{[3-(TRIFLUOROMETHYL)PHENYL]AMINO}NICOTINIC ACID, ... | | 著者 | Capper, M.J, Mathiharan, Y.K, Yang, S, Stone, A.C, Wacker, D. | | 登録日 | 2022-02-11 | | 公開日 | 2023-08-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | Substrate binding and inhibition of the anion exchanger 1 transporter.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6WDP
 
 | | Interleukin 12 receptor subunit beta-1 | | 分子名称: | GLYCEROL, Interleukin-12 receptor subunit beta-1, SULFATE ION | | 著者 | Spangler, J.B, Thomas, C, Jude, K.M, Garcia, K.C. | | 登録日 | 2020-04-01 | | 公開日 | 2021-02-24 | | 最終更新日 | 2021-12-01 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Structural basis for IL-12 and IL-23 receptor sharing reveals a gateway for shaping actions on T versus NK cells.
Cell, 184, 2021
|
|
6WDQ
 
 | | IL23/IL23R/IL12Rb1 signaling complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-12 receptor subunit beta-1, ... | | 著者 | Jude, K.M, Ely, L.K, Glassman, C.R, Thomas, C, Spangler, J.B, Lupardus, P.J, Garcia, K.C. | | 登録日 | 2020-04-01 | | 公開日 | 2021-02-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Structural basis for IL-12 and IL-23 receptor sharing reveals a gateway for shaping actions on T versus NK cells.
Cell, 184, 2021
|
|
