1KP6
 
 | | USTILAGO MAYDIS KILLER TOXIN KP6 ALPHA-SUBUNIT | | 分子名称: | PROTEIN (TOXIN), SULFATE ION | | 著者 | Li, N, Erman, M, Pangborn, W, Duax, W.L, Park, C.-M, Bruenn, J, Ghosh, D. | | 登録日 | 1999-05-28 | | 公開日 | 1999-07-21 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure of Ustilago maydis killer toxin KP6 alpha-subunit. A multimeric assembly with a central pore.
J.Biol.Chem., 274, 1999
|
|
3J3W
 
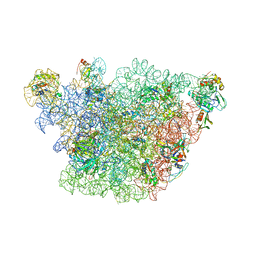 | | Atomic model of the immature 50S subunit from Bacillus subtilis (state II-a) | | 分子名称: | 50S ribosomal protein L1, 50S ribosomal protein L11, 50S ribosomal protein L13, ... | | 著者 | Li, N, Guo, Q, Zhang, Y, Yuan, Y, Ma, C, Lei, J, Gao, N. | | 登録日 | 2013-04-28 | | 公開日 | 2013-06-12 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (10.7 Å) | | 主引用文献 | Cryo-EM structures of the late-stage assembly intermediates of the bacterial 50S ribosomal subunit
Nucleic Acids Res., 41, 2013
|
|
3J3V
 
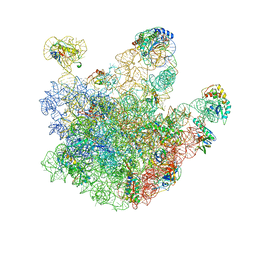 | | Atomic model of the immature 50S subunit from Bacillus subtilis (state I-a) | | 分子名称: | 50S ribosomal protein L1, 50S ribosomal protein L11, 50S ribosomal protein L13, ... | | 著者 | Li, N, Guo, Q, Zhang, Y, Yuan, Y, Ma, C, Lei, J, Gao, N. | | 登録日 | 2013-04-28 | | 公開日 | 2013-06-12 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (13.3 Å) | | 主引用文献 | Cryo-EM structures of the late-stage assembly intermediates of the bacterial 50S ribosomal subunit
Nucleic Acids Res., 41, 2013
|
|
7FGF
 
 | |
8XGC
 
 | | Structure of yeast replisome associated with FACT and histone hexamer, Composite map | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45, Chromosome segregation in meiosis protein 3, ... | | 著者 | Li, N, Gao, Y, Yu, D, Gao, N, Zhai, Y. | | 登録日 | 2023-12-15 | | 公開日 | 2024-02-14 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Parental histone transfer caught at the replication fork.
Nature, 627, 2024
|
|
3JA8
 
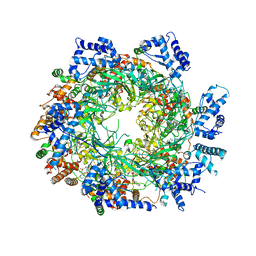 | | Cryo-EM structure of the MCM2-7 double hexamer | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Minichromosome Maintenance 2, Minichromosome Maintenance 3, ... | | 著者 | Li, N, Zhai, Y, Zhang, Y, Li, W, Yang, M, Lei, J, Tye, B.K, Gao, N. | | 登録日 | 2015-05-09 | | 公開日 | 2015-08-05 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structure of the eukaryotic MCM complex at 3.8 angstrom
Nature, 524, 2015
|
|
3M1H
 
 | |
1I1S
 
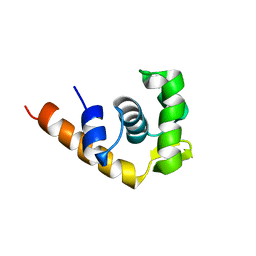 | | SOLUTION STRUCTURE OF THE TRANSCRIPTIONAL ACTIVATION DOMAIN OF THE BACTERIOPHAGE T4 PROTEIN MOTA | | 分子名称: | MOTA | | 著者 | Li, N, Zhang, W, White, S.W, Kriwacki, R.W. | | 登録日 | 2001-02-02 | | 公開日 | 2001-02-14 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the transcriptional activation domain of the bacteriophage T4 protein, MotA.
Biochemistry, 40, 2001
|
|
5WUA
 
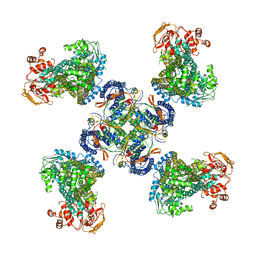 | | Structure of a Pancreatic ATP-sensitive Potassium Channel | | 分子名称: | ATP-sensitive inward rectifier potassium channel 11,superfolder GFP, SUR1 | | 著者 | Li, N, Wu, J.-X, Chen, L, Gao, N. | | 登録日 | 2016-12-16 | | 公開日 | 2017-01-25 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (5.6 Å) | | 主引用文献 | Structure of a Pancreatic ATP-Sensitive Potassium Channel
Cell, 168, 2017
|
|
1KAF
 
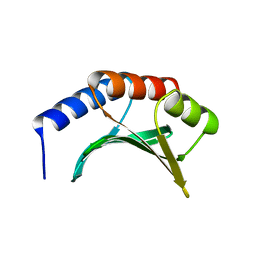 | | DNA Binding Domain Of The Phage T4 Transcription Factor MotA (AA105-211) | | 分子名称: | Transcription regulatory protein MOTA | | 著者 | Li, N, Sickmier, E.A, Zhang, R, Joachimiak, A, White, S.W. | | 登録日 | 2001-11-01 | | 公開日 | 2001-11-21 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The MotA transcription factor from bacteriophage T4 contains a novel DNA-binding domain: the 'double wing' motif.
Mol.Microbiol., 43, 2002
|
|
8IB2
 
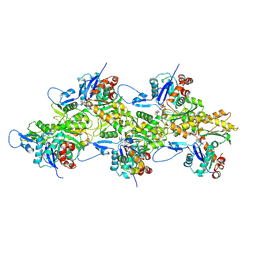 | | Structure of mammalian spectrin-actin junctional complex of membrane skeleton, Pointed-end segment, headpiece domain of dematin optimized | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | 著者 | Li, N, Chen, S, Gao, N. | | 登録日 | 2023-02-09 | | 公開日 | 2023-04-26 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural basis of membrane skeleton organization in red blood cells.
Cell, 186, 2023
|
|
8IAH
 
 | | Structure of mammalian spectrin-actin junctional complex of membrane skeleton, State I, Global map | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | 著者 | Li, N, Chen, S, Gao, N. | | 登録日 | 2023-02-08 | | 公開日 | 2023-05-03 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural basis of membrane skeleton organization in red blood cells.
Cell, 186, 2023
|
|
8IAI
 
 | | Structure of mammalian spectrin-actin junctional complex of membrane skeleton, State II, Global map | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | 著者 | Li, N, Chen, S, Gao, N. | | 登録日 | 2023-02-08 | | 公開日 | 2023-05-03 | | 最終更新日 | 2025-06-18 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural basis of membrane skeleton organization in red blood cells.
Cell, 186, 2023
|
|
6P7B
 
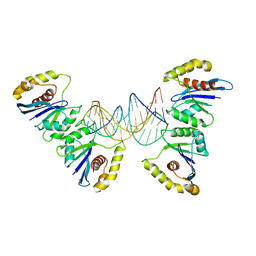 | | Crystal structure of Fowlpox virus resolvase and substrate Holliday junction DNA complex | | 分子名称: | DNA (29-MER), Holliday junction resolvase | | 著者 | Li, N, Shi, K, Rao, T, Banerjee, S, Aihara, H. | | 登録日 | 2019-06-05 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3.317 Å) | | 主引用文献 | Structural insights into the promiscuous DNA binding and broad substrate selectivity of fowlpox virus resolvase.
Sci Rep, 10, 2020
|
|
6P7A
 
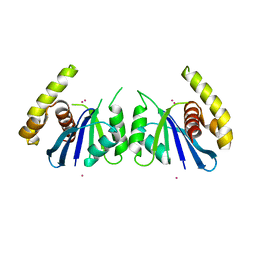 | | CRYSTAL STRUCTURE OF THE FOWLPOX VIRUS HOLLIDAY JUNCTION RESOLVASE | | 分子名称: | CADMIUM ION, Holliday junction resolvase | | 著者 | Li, N, Shi, K, Banerjee, S, Rao, T, Aihara, H. | | 登録日 | 2019-06-05 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3.081 Å) | | 主引用文献 | Structural insights into the promiscuous DNA binding and broad substrate selectivity of fowlpox virus resolvase.
Sci Rep, 10, 2020
|
|
3R5T
 
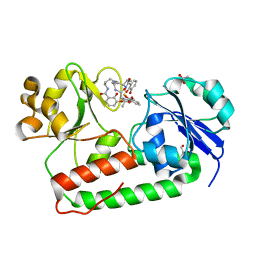 | | Crystal structure of holo-ViuP | | 分子名称: | (4S,5R)-N-{3-[(2,3-dihydroxybenzoyl)amino]propyl}-2-(2,3-dihydroxyphenyl)-N-[3-({[(4S,5R)-2-(2,3-dihydroxyphenyl)-5-met hyl-4,5-dihydro-1,3-oxazol-4-yl]carbonyl}amino)propyl]-5-methyl-4,5-dihydro-1,3-oxazole-4-carboxamide, 1,2-ETHANEDIOL, ACETIC ACID, ... | | 著者 | Li, N, Zhang, C, Li, B, Liu, X, Huang, Y, Xu, S, Gu, L. | | 登録日 | 2011-03-19 | | 公開日 | 2012-02-08 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Unique iron coordination in iron-chelating molecule vibriobactin helps Vibrio cholerae evade mammalian siderocalin-mediated immune response.
J.Biol.Chem., 287, 2012
|
|
3R5S
 
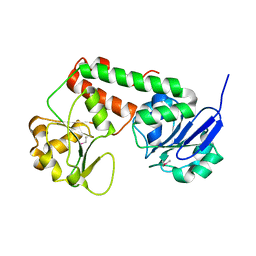 | | Crystal structure of apo-ViuP | | 分子名称: | Ferric vibriobactin ABC transporter, periplasmic ferric vibriobactin-binding protein | | 著者 | Li, N, Zhang, C, Li, B, Liu, X, Huang, Y, Xu, S, Gu, L. | | 登録日 | 2011-03-19 | | 公開日 | 2012-02-08 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.791 Å) | | 主引用文献 | Unique iron coordination in iron-chelating molecule vibriobactin helps Vibrio cholerae evade mammalian siderocalin-mediated immune response.
J.Biol.Chem., 287, 2012
|
|
5ZR1
 
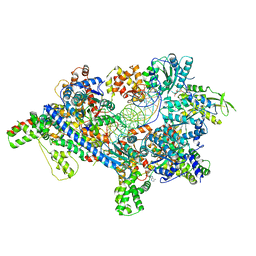 | | Saccharomyces Cerevisiae Origin Recognition Complex Bound to a 72-bp Origin DNA containing ACS and B1 element | | 分子名称: | 72bp-oring DNA, ACS305, A-rich, ... | | 著者 | Li, N, Lam, W.H, Zhai, Y, Cheng, J, Cheng, E, Zhao, Y, Gao, N, Tye, B.K. | | 登録日 | 2018-04-21 | | 公開日 | 2018-07-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structure of the origin recognition complex bound to DNA replication origin.
Nature, 559, 2018
|
|
3KM5
 
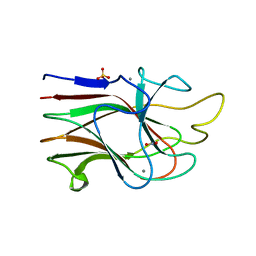 | | Crystal Structure Analysis of the K2 Cleaved Adhesin Domain of Lys-gingipain (Kgp) | | 分子名称: | CALCIUM ION, GLYCEROL, Lysine specific cysteine protease, ... | | 著者 | Li, N, Collyer, C.A, Hunter, N. | | 登録日 | 2009-11-09 | | 公開日 | 2010-03-31 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structure determination and analysis of a haemolytic gingipain adhesin domain from Porphyromonas gingivalis
Mol.Microbiol., 76, 2010
|
|
7F77
 
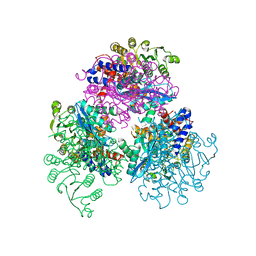 | | Crystal structure of glutamate dehydrogenase 3 from Candida albicans | | 分子名称: | Glutamate dehydrogenase | | 著者 | Li, N, Wang, W, Zeng, X, Liu, M, Li, M, Li, C, Wang, M. | | 登録日 | 2021-06-28 | | 公開日 | 2021-07-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.086 Å) | | 主引用文献 | Crystal structure of glutamate dehydrogenase 3 from Candida albicans.
Biochem.Biophys.Res.Commun., 570, 2021
|
|
7F79
 
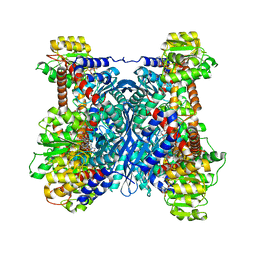 | | Crystal structure of glutamate dehydrogenase 3 from Candida albicans in complex with alpha-ketoglutarate and NADPH | | 分子名称: | 2-OXOGLUTARIC ACID, GLYCEROL, Glutamate dehydrogenase, ... | | 著者 | Li, N, Wang, W, Zeng, X, Liu, M, Li, M, Li, C, Wang, M. | | 登録日 | 2021-06-28 | | 公開日 | 2021-07-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure of glutamate dehydrogenase 3 from Candida albicans.
Biochem.Biophys.Res.Commun., 570, 2021
|
|
8J59
 
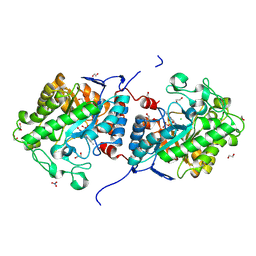 | | The structure of a novel thermophilic-like old yellow enzyme from Aspergillus flavus-AfOYE1 | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | 著者 | Li, N, Wang, Y. | | 登録日 | 2023-04-21 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structural and functional characterization of a new thermophilic-like OYE from Aspergillus flavus.
Appl.Microbiol.Biotechnol., 108, 2024
|
|
8X0T
 
 | | Frizzled8 CRD in complex with pF8_AC3 Fab | | 分子名称: | Frizzled-8, pF8_AC3 Heavy chain, pF8_AC3 Light chain | | 著者 | Li, N, Ge, Q. | | 登録日 | 2023-11-06 | | 公開日 | 2023-11-22 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Identification and functional validation of FZD8-specific antibodies.
Int J Biol Macromol, 254, 2023
|
|
3TLK
 
 | | Crystal structure of holo FepB | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, FE (III) ION, Ferrienterobactin-binding periplasmic protein, ... | | 著者 | Li, N, Gu, L. | | 登録日 | 2011-08-30 | | 公開日 | 2013-01-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.853 Å) | | 主引用文献 | Crystal structure of holo FepB
To be Published
|
|
1M53
 
 | |
