3WZ8
 
 | | Endothiapepsin in complex with Gewald reaction-derived inhibitor (8) | | 分子名称: | DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | 著者 | Kuhnert, M, Steuber, H, Diederich, W.E. | | 登録日 | 2014-09-19 | | 公開日 | 2015-08-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Tracing binding modes in hit-to-lead optimization: chameleon-like poses of aspartic protease inhibitors
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
3WZ6
 
 | | Endothiapepsin in complex with Gewald reaction-derived inhibitor (5) | | 分子名称: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Kuhnert, M, Steuber, H, Diederich, W.E. | | 登録日 | 2014-09-19 | | 公開日 | 2015-08-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.404 Å) | | 主引用文献 | Tracing binding modes in hit-to-lead optimization: chameleon-like poses of aspartic protease inhibitors
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
3WZ7
 
 | | Endothiapepsin in complex with Gewald reaction-derived inhibitor (6) | | 分子名称: | DIMETHYL SULFOXIDE, Endothiapepsin, N-benzyl-2-({N-[2-(1H-indol-3-yl)ethyl]glycyl}amino)-4,5,6,7-tetrahydro-1-benzothiophene-3-carboxamide | | 著者 | Kuhnert, M, Steuber, H, Diederich, W.E. | | 登録日 | 2014-09-19 | | 公開日 | 2015-08-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Tracing binding modes in hit-to-lead optimization: chameleon-like poses of aspartic protease inhibitors
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4YDG
 
 | | Crystal structure of compound 10 in complex with HTLV-1 Protease | | 分子名称: | HTLV-1 protease, N,N'-(3S,4S)-PYRROLIDINE-3,4-DIYLBIS(4-AMINO-N-BENZYLBENZENESULFONAMIDE), SULFATE ION | | 著者 | Kuhnert, M, Blum, A, Steuber, H, Diederich, W.E. | | 登録日 | 2015-02-22 | | 公開日 | 2016-02-03 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | Privileged Structures Meet Human T-Cell Leukemia Virus-1 (HTLV-1): C2-Symmetric 3,4-Disubstituted Pyrrolidines as Nonpeptidic HTLV-1 Protease Inhibitors.
J.Med.Chem., 58, 2015
|
|
4YDF
 
 | | Crystal structure of compound 9 in complex with HTLV-1 Protease | | 分子名称: | HTLV-1 Protease, N-benzyl-N-[(3S,4S)-4-{benzyl[(4-nitrophenyl)sulfonyl]amino}pyrrolidin-3-yl]-3-nitrobenzenesulfonamide, SULFATE ION | | 著者 | Kuhnert, M, Blum, A, Steuber, H, Diederich, W.E. | | 登録日 | 2015-02-22 | | 公開日 | 2016-02-03 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.804 Å) | | 主引用文献 | Privileged Structures Meet Human T-Cell Leukemia Virus-1 (HTLV-1): C2-Symmetric 3,4-Disubstituted Pyrrolidines as Nonpeptidic HTLV-1 Protease Inhibitors.
J.Med.Chem., 58, 2015
|
|
3WSJ
 
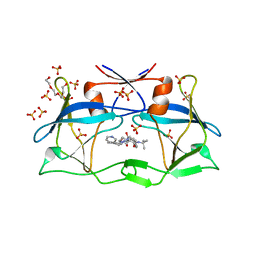 | | HTLV-1 protease in complex with the HIV-1 protease inhibitor Indinavir | | 分子名称: | N-[2(R)-HYDROXY-1(S)-INDANYL]-5-[(2(S)-TERTIARY BUTYLAMINOCARBONYL)-4(3-PYRIDYLMETHYL)PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYLPENTANAMIDE, Protease, SULFATE ION, ... | | 著者 | Kuhnert, M, Steuber, H, Diederich, W.E. | | 登録日 | 2014-03-14 | | 公開日 | 2014-10-22 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.404 Å) | | 主引用文献 | Structural basis for HTLV-1 protease inhibition by the HIV-1 protease inhibitor indinavir.
J.Med.Chem., 57, 2014
|
|
4RS2
 
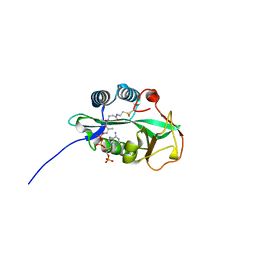 | | 1.55 Angstrom Crystal Structure of GNAT Family N-acetyltransferase (YhbS) from Escherichia coli in Complex with CoA | | 分子名称: | COENZYME A, Predicted acyltransferase with acyl-CoA N-acyltransferase domain | | 著者 | Minasov, G, Wawrzak, Z, Kuhn, M, Shuvalova, L, Dubrovska, I, Flores, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2014-11-06 | | 公開日 | 2014-11-19 | | 最終更新日 | 2017-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | 1.55 Angstrom Crystal Structure of GNAT Family N-acetyltransferase (YhbS) from Escherichia coli in Complex with CoA.
TO BE PUBLISHED
|
|
3T41
 
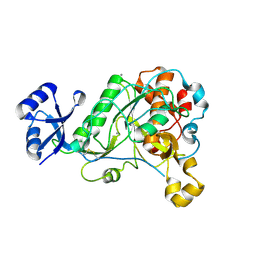 | | 1.95 Angstrom Resolution Crystal Structure of Epidermin Leader Peptide Processing Serine Protease (EpiP) S393A Mutant from Staphylococcus aureus | | 分子名称: | CALCIUM ION, CHLORIDE ION, Epidermin leader peptide processing serine protease EpiP | | 著者 | Minasov, G, Kuhn, M, Ruan, J, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-07-25 | | 公開日 | 2011-08-17 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | 1.95 Angstrom Resolution Crystal Structure of Epidermin Leader Peptide Processing Serine Protease (EpiP) S393A Mutant from Staphylococcus aureus.
TO BE PUBLISHED
|
|
3UWQ
 
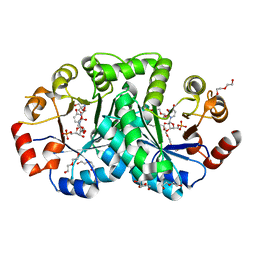 | | 1.80 Angstrom resolution crystal structure of orotidine 5'-phosphate decarboxylase from Vibrio cholerae O1 biovar eltor str. N16961 in complex with uridine-5'-monophosphate (UMP) | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Halavaty, A.S, Minasov, G, Winsor, J, Shuvalova, L, Kuhn, M, Filippova, E.V, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-12-02 | | 公開日 | 2011-12-14 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | 1.80 Angstrom resolution crystal structure of orotidine 5'-phosphate decarboxylase from Vibrio cholerae O1 biovar eltor str. N16961 in complex with uridine-5'-monophosphate (UMP)
To be Published
|
|
4FCE
 
 | | Crystal structure of Yersinia pestis GlmU in complex with alpha-D-glucosamine 1-phosphate (GP1) | | 分子名称: | 1,2-ETHANEDIOL, 2-amino-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, Bifunctional protein GlmU, ... | | 著者 | Nocek, B, Kuhn, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2012-05-24 | | 公開日 | 2012-07-11 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.955 Å) | | 主引用文献 | Crystal structure of Yersinia pestis GlmU in complex with alpha-D-glucosamine 1-phosphate (GP1)
To be Published
|
|
6PXA
 
 | | The crystal structure of chloramphenicol acetyltransferase-like protein from Vibrio fischeri ES114 in complex with taurocholic acid | | 分子名称: | ACETATE ION, CHLORIDE ION, Chloramphenicol acetyltransferase, ... | | 著者 | Tan, K, Maltseva, N, Jedrzejczak, R, Kuhn, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-07-25 | | 公開日 | 2019-09-25 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | The crystal structure of chloramphenicol acetyltransferase-like protein from Vibrio fischeri ES114 in complex with taurocholic acid
To Be Published
|
|
6PUB
 
 | | Crystal Structure of the Type B Chloramphenicol Acetyltransferase from Vibrio cholerae in the Complex with Crystal Violet | | 分子名称: | CHLORIDE ION, CRYSTAL VIOLET, Chloramphenicol acetyltransferase, ... | | 著者 | Kim, Y, Maltseva, N, Kuhn, M, Stam, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-07-18 | | 公開日 | 2019-09-25 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.43 Å) | | 主引用文献 | Crystal Structure of the Type B Chloramphenicol Acetyltransferase from Vibrio cholerae in the Complex with Crystal Violet
To Be Published
|
|
4L6B
 
 | |
4LAP
 
 | |
3PSY
 
 | | Endothiapepsin in complex with an inhibitor based on the Gewald reaction | | 分子名称: | DIMETHYL SULFOXIDE, Endothiapepsin, GLYCEROL, ... | | 著者 | Koester, H, Heine, A, Klebe, G. | | 登録日 | 2010-12-02 | | 公開日 | 2011-12-07 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | Tracing binding modes in hit-to-lead optimization: chameleon-like poses of aspartic protease inhibitors.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6PFY
 
 | |
6PGK
 
 | | Membrane Protein Megahertz Crystallography at the European XFEL, Photosystem I XFEL at 2.9 A | | 分子名称: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | 著者 | Fromme, R, Gisriel, C, Fromme, P. | | 登録日 | 2019-06-24 | | 公開日 | 2019-11-27 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Membrane protein megahertz crystallography at the European XFEL.
Nat Commun, 10, 2019
|
|
6BI4
 
 | | 2.9 Angstrom Resolution Crystal Structure of dTDP-Glucose 4,6-dehydratase (rfbB) from Bacillus anthracis str. Ames in Complex with NAD. | | 分子名称: | NICKEL (II) ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | 著者 | Halavaty, A.S, Kuhn, M, Shuvalova, L, Minasov, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-10-31 | | 公開日 | 2017-11-08 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.91 Å) | | 主引用文献 | Structure of the Bacillus anthracis dTDP-l-rhamnose biosynthetic pathway enzyme: dTDP-alpha-d-glucose 4,6-dehydratase, RfbB.
J.Struct.Biol., 202, 2018
|
|
8A1I
 
 | |
6WEB
 
 | | Multi-Hit SFX using MHz XFEL sources | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | 著者 | Holmes, S, Darmanin, C, Abbey, B. | | 登録日 | 2020-04-01 | | 公開日 | 2021-10-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Megahertz pulse trains enable multi-hit serial femtosecond crystallography experiments at X-ray free electron lasers.
Nat Commun, 13, 2022
|
|
6WEC
 
 | | Multi-Hit SFX using MHz XFEL sources | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | 著者 | Holmes, S, Darmanin, C, Abbey, B. | | 登録日 | 2020-04-01 | | 公開日 | 2021-10-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Megahertz pulse trains enable multi-hit serial femtosecond crystallography experiments at X-ray free electron lasers.
Nat Commun, 13, 2022
|
|
7PVN
 
 | | Crystal Structure of Human UBA6 in Complex with ATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Truongvan, N, Li, S, Schindelin, H. | | 登録日 | 2021-10-05 | | 公開日 | 2022-08-31 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | Structures of UBA6 explain its dual specificity for ubiquitin and FAT10.
Nat Commun, 13, 2022
|
|
4MVJ
 
 | | 2.85 Angstrom Resolution Crystal Structure of Glyceraldehyde 3-phosphate Dehydrogenase A (gapA) from Escherichia coli Modified by Acetyl Phosphate. | | 分子名称: | ACETATE ION, ACETYLPHOSPHATE, CHLORIDE ION, ... | | 著者 | Minasov, G, Kuhn, M, Dubrovska, I, Winsor, J, Shuvalova, L, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-09-24 | | 公開日 | 2014-04-23 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Structural, kinetic and proteomic characterization of acetyl phosphate-dependent bacterial protein acetylation.
Plos One, 9, 2014
|
|
7PYV
 
 | | Crystal structure of human UBA6 in complex with the ubiquitin-like modifier FAT10 | | 分子名称: | UBD, Ubiquitin-like modifier-activating enzyme 6,Ubiquitin-like modifier-activating enzyme 1,Ubiquitin-like modifier-activating enzyme 6 | | 著者 | Li, S, Truongvan, N, Schindelin, H. | | 登録日 | 2021-10-11 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.27 Å) | | 主引用文献 | Structures of UBA6 explain its dual specificity for ubiquitin and FAT10.
Nat Commun, 13, 2022
|
|
4K6A
 
 | | Revised Crystal Structure of apo-form of Triosephosphate Isomerase (tpiA) from Escherichia coli at 1.8 Angstrom Resolution. | | 分子名称: | SODIUM ION, Triosephosphate isomerase | | 著者 | Minasov, G, Kuhn, M, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Grimshaw, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-04-15 | | 公開日 | 2013-05-01 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural, kinetic and proteomic characterization of acetyl phosphate-dependent bacterial protein acetylation.
PLoS ONE, 9, 2014
|
|
