6HQA
 
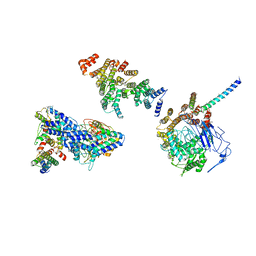 | | Molecular structure of promoter-bound yeast TFIID | | 分子名称: | Histone-fold, Subunit (17 kDa) of TFIID and SAGA complexes, involved in RNA polymerase II transcription initiation, ... | | 著者 | Kolesnikova, O, Ben-Shem, A, Luo, J, Ranish, J, Schultz, P, Papai, G. | | 登録日 | 2018-09-24 | | 公開日 | 2018-11-21 | | 最終更新日 | 2022-03-30 | | 実験手法 | ELECTRON MICROSCOPY (7.1 Å) | | 主引用文献 | Molecular structure of promoter-bound yeast TFIID.
Nat Commun, 9, 2018
|
|
6TBM
 
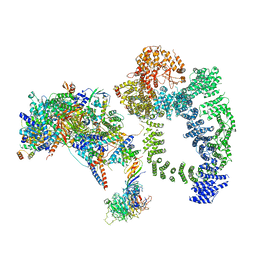 | | Structure of SAGA bound to TBP, including Spt8 and DUB | | 分子名称: | Polyubiquitin-B, SAGA-associated factor 11, Spt20, ... | | 著者 | Papai, G, Frechard, A, Kolesnikova, O, Crucifix, C, Schultz, P, Ben-Shem, A. | | 登録日 | 2019-11-01 | | 公開日 | 2020-02-12 | | 最終更新日 | 2021-06-30 | | 実験手法 | ELECTRON MICROSCOPY (20 Å) | | 主引用文献 | Structure of SAGA and mechanism of TBP deposition on gene promoters.
Nature, 577, 2020
|
|
6TB4
 
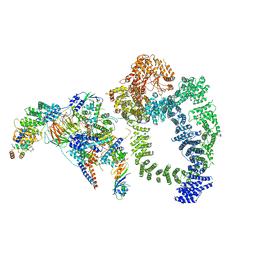 | | Structure of SAGA bound to TBP | | 分子名称: | SAGA-associated factor 73 (Sgf73), Spt20, Subunit (17 kDa) of TFIID and SAGA complexes, ... | | 著者 | Papai, G, Frechard, A, Kolesnikova, O, Crucifix, C, Schultz, P, Ben-Shem, A. | | 登録日 | 2019-10-31 | | 公開日 | 2020-01-29 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structure of SAGA and mechanism of TBP deposition on gene promoters.
Nature, 577, 2020
|
|
4OGP
 
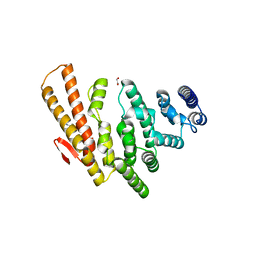 | | Structure of C-terminal domain from S. cerevisiae Pat1 decapping activator (Space group : P21) | | 分子名称: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA topoisomerase 2-associated protein PAT1 | | 著者 | Fourati-Kammoun, Z, Kolesnikova, O, Back, R, Keller, J, Lazar, N, Gaudon-Plesse, C, Seraphin, B, Graille, M. | | 登録日 | 2014-01-16 | | 公開日 | 2014-10-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | The C-terminal domain from S. cerevisiae Pat1 displays two conserved regions involved in decapping factor recruitment.
Plos One, 9, 2014
|
|
4OJJ
 
 | | Structure of C-terminal domain from S. cerevisiae Pat1 decapping activator (Space group : P212121) | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA topoisomerase 2-associated protein PAT1, ... | | 著者 | Fourati-Kammoun, Z, Kolesnikova, O, Back, R, Keller, J, Lazar, N, Gaudon-Plesse, C, Seraphin, B, Graille, M. | | 登録日 | 2014-01-21 | | 公開日 | 2014-10-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | The C-terminal domain from S. cerevisiae Pat1 displays two conserved regions involved in decapping factor recruitment.
Plos One, 9, 2014
|
|
5LM5
 
 | | Structure of C-terminal domain from S. cerevisiae Pat1 decapping activator bound to Dcp2 HLM2 peptide (region 435-451) | | 分子名称: | DNA topoisomerase 2-associated protein PAT1, mRNA decapping protein 2 | | 著者 | Charenton, C, Gaudon-Plesse, C, Fourati, Z, Taverniti, V, Back, R, Kolesnikova, O, Seraphin, B, Graille, M. | | 登録日 | 2016-07-29 | | 公開日 | 2017-08-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | A unique surface on Pat1 C-terminal domain directly interacts with Dcp2 decapping enzyme and Xrn1 5'-3' mRNA exonuclease in yeast.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5LMG
 
 | | Structure of C-terminal domain from S. cerevisiae Pat1 decapping activator bound to Dcp2 HLM10 peptide (region 954-970) | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA topoisomerase 2-associated protein PAT1, ... | | 著者 | Charenton, C, Gaudon-Plesse, C, Fourati, Z, Taverniti, V, Back, R, Kolesnikova, O, Seraphin, B, Graille, M. | | 登録日 | 2016-07-30 | | 公開日 | 2017-08-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | A unique surface on Pat1 C-terminal domain directly interacts with Dcp2 decapping enzyme and Xrn1 5'-3' mRNA exonuclease in yeast.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5LMF
 
 | | Structure of C-terminal domain from S. cerevisiae Pat1 decapping activator bound to Dcp2 HLM3 peptide (region 484-500) | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA topoisomerase 2-associated protein PAT1, ... | | 著者 | Charenton, C, Gaudon-Plesse, C, Fourati, Z, Taverniti, V, Back, R, Kolesnikova, O, Seraphin, B, Graille, M. | | 登録日 | 2016-07-30 | | 公開日 | 2017-08-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | A unique surface on Pat1 C-terminal domain directly interacts with Dcp2 decapping enzyme and Xrn1 5'-3' mRNA exonuclease in yeast.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5OEJ
 
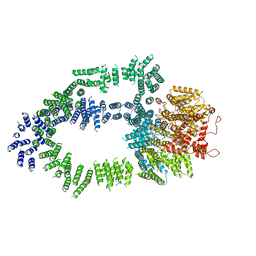 | | Structure of Tra1 subunit within the chromatin modifying complex SAGA | | 分子名称: | Tra1 subunit within the chromatin modifying complex SAGA | | 著者 | Sharov, G, Voltz, K, Durand, A, Kolesnikova, O, Papai, G, Myasnikov, A.G, Dejaegere, A, Ben-Shem, A, Schultz, P. | | 登録日 | 2017-07-07 | | 公開日 | 2017-08-02 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (5.7 Å) | | 主引用文献 | Structure of the transcription activator target Tra1 within the chromatin modifying complex SAGA.
Nat Commun, 8, 2017
|
|
8OO7
 
 | | CryoEM Structure INO80core Hexasome complex composite model state1 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 5, ... | | 著者 | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | 登録日 | 2023-04-04 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOP
 
 | | CryoEM Structure INO80core Hexasome complex composite model state2 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 5, ... | | 著者 | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | 登録日 | 2023-04-05 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOR
 
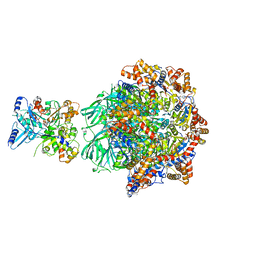 | | CryoEM Structure INO80core Hexasome complex Rvb core refinement state2 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 5, ... | | 著者 | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | 登録日 | 2023-04-05 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.87 Å) | | 主引用文献 | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOK
 
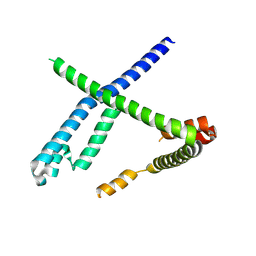 | |
8OOF
 
 | | CryoEM Structure INO80core Hexasome complex Arp5 Ies6 refinement state1 | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling complex subunit IES6, ... | | 著者 | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | 登録日 | 2023-04-05 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OO9
 
 | | CryoEM Structure INO80core Hexasome complex ATPase-DNA refinement state1 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Chromatin-remodeling ATPase INO80, DNA strand 1, ... | | 著者 | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | 登録日 | 2023-04-04 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOT
 
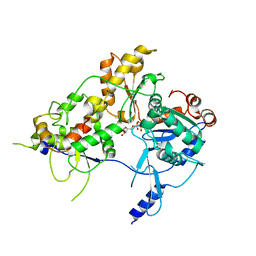 | | CryoEM Structure INO80core Hexasome complex Arp5 Ies6 refinement state2 | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling complex subunit IES6, ... | | 著者 | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | 登録日 | 2023-04-05 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOA
 
 | | CryoEM Structure INO80core Hexasome complex Hexasome refinement state1 | | 分子名称: | DNA Strand 2, DNA strand 1, Histone H2A, ... | | 著者 | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | 登録日 | 2023-04-04 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOS
 
 | | CryoEM Structure INO80core Hexasome complex ATPase-hexasome refinement state 2 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Chromatin-remodeling ATPase Ino80, DNA Strand 2, ... | | 著者 | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | 登録日 | 2023-04-05 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.29 Å) | | 主引用文献 | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OOC
 
 | | CryoEM Structure INO80core Hexasome complex Rvb core refinement state1 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Chromatin-remodeling ATPase Ino80, ... | | 著者 | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | 登録日 | 2023-04-05 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.93 Å) | | 主引用文献 | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
