5VKQ
 
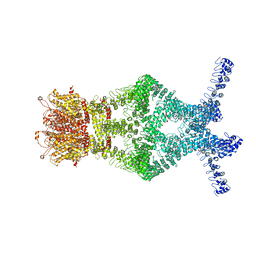 | | Structure of a mechanotransduction ion channel Drosophila NOMPC in nanodisc | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, No mechanoreceptor potential C isoform L | | 著者 | Jin, P, Bulkley, D, Guo, Y, Zhang, W, Guo, Z, Huynh, W, Wu, S, Meltzer, S, Chen, T, Jan, L.Y, Jan, Y.-N, Cheng, Y. | | 登録日 | 2017-04-22 | | 公開日 | 2017-06-28 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.55 Å) | | 主引用文献 | Electron cryo-microscopy structure of the mechanotransduction channel NOMPC.
Nature, 547, 2017
|
|
6P49
 
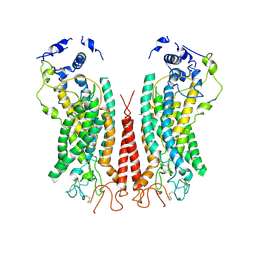 | | Cryo-EM structure of calcium-bound TMEM16F in nanodisc with supplement of PIP2 in Cl2 | | 分子名称: | Anoctamin-6, CALCIUM ION | | 著者 | Feng, S, Dang, S, Han, T.W, Ye, W, Jin, P, Cheng, T, Li, J, Jan, Y.N, Jan, L.Y, Cheng, Y. | | 登録日 | 2019-05-26 | | 公開日 | 2019-07-24 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Cryo-EM Studies of TMEM16F Calcium-Activated Ion Channel Suggest Features Important for Lipid Scrambling.
Cell Rep, 28, 2019
|
|
6P46
 
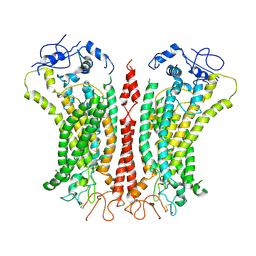 | | Cryo-EM structure of TMEM16F in digitonin with calcium bound | | 分子名称: | Anoctamin-6, CALCIUM ION | | 著者 | Feng, S, Dang, S, Han, T.W, Ye, W, Jin, P, Cheng, T, Li, J, Jan, Y.N, Jan, L.Y, Cheng, Y. | | 登録日 | 2019-05-26 | | 公開日 | 2019-07-24 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Cryo-EM Studies of TMEM16F Calcium-Activated Ion Channel Suggest Features Important for Lipid Scrambling.
Cell Rep, 28, 2019
|
|
6P47
 
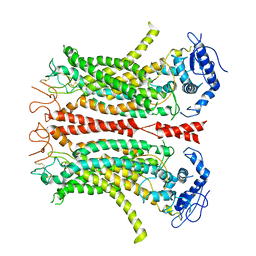 | | Cryo-EM structure of TMEM16F in digitonin without calcium | | 分子名称: | Anoctamin-6 | | 著者 | Feng, S, Dang, S, Han, T.W, Ye, W, Jin, P, Cheng, T, Li, J, Jan, Y.N, Jan, L.Y, Cheng, Y. | | 登録日 | 2019-05-26 | | 公開日 | 2019-07-24 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Cryo-EM Studies of TMEM16F Calcium-Activated Ion Channel Suggest Features Important for Lipid Scrambling.
Cell Rep, 28, 2019
|
|
6P48
 
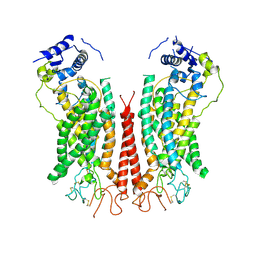 | | Cryo-EM structure of calcium-bound TMEM16F in nanodisc with supplement of PIP2 in Cl1 | | 分子名称: | Anoctamin-6, CALCIUM ION | | 著者 | Feng, S, Dang, S, Han, T.W, Ye, W, Jin, P, Cheng, T, Li, J, Jan, Y.N, Jan, L.Y, Cheng, Y. | | 登録日 | 2019-05-26 | | 公開日 | 2019-07-24 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM Studies of TMEM16F Calcium-Activated Ion Channel Suggest Features Important for Lipid Scrambling.
Cell Rep, 28, 2019
|
|
2ASK
 
 | | Structure of human Artemin | | 分子名称: | SULFATE ION, artemin | | 著者 | Silvian, L, Jin, P, Carmillo, P, Boriack-Sjodin, P.A, Pelletier, C, Rushe, M, Gong, B.J, Sah, D, Pepinsky, B, Rossomando, A. | | 登録日 | 2005-08-23 | | 公開日 | 2006-06-13 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Artemin crystal structure reveals insights into heparan sulfate binding.
Biochemistry, 45, 2006
|
|
5KE9
 
 | |
5KEA
 
 | |
7BPV
 
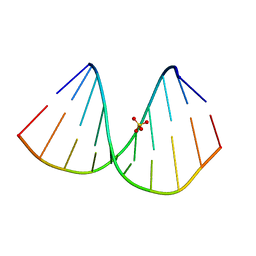 | | r(GUGGGCCGAC)/d(GTCGGCCCAC) hybrid duplex structure | | 分子名称: | DNA (5'-D(P*GP*TP*CP*GP*GP*CP*CP*CP*AP*C)-3'), RNA (5'-R(P*GP*UP*GP*GP*GP*CP*CP*GP*AP*C)-3'), SULFATE ION | | 著者 | Wang, S.C, Satange, R.B, Hou, M.H. | | 登録日 | 2020-03-23 | | 公開日 | 2021-03-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.781 Å) | | 主引用文献 | Targeting the ALS/FTD-associated A-DNA kink with anthracene-based metal complex causes DNA backbone straightening and groove contraction.
Nucleic Acids Res., 49, 2021
|
|
6W46
 
 | |
6W47
 
 | |
5HHX
 
 | |
5KE7
 
 | |
5KE6
 
 | |
5KEB
 
 | |
5HHV
 
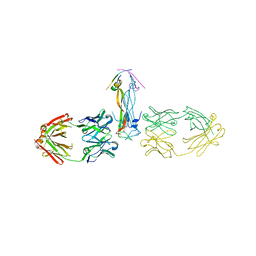 | |
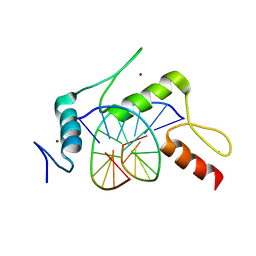
5KE8
 
 | |
4GPI
 
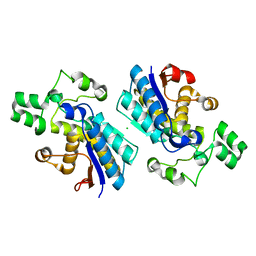 | |
4GPZ
 
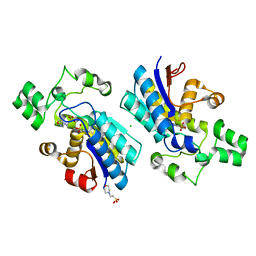 | |
4IN4
 
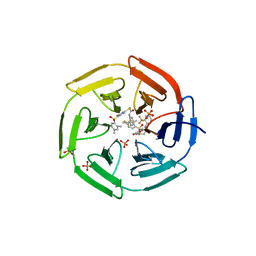 | | Crystal structure of cpd 15 bound to Keap1 Kelch domain | | 分子名称: | 2-({5-[(2,4-dimethylphenyl)sulfonyl]-6-oxo-1,6-dihydropyrimidin-2-yl}sulfanyl)-N-[2-(trifluoromethyl)phenyl]acetamide, Kelch-like ECH-associated protein 1, PHOSPHATE ION | | 著者 | Silvian, L, Marcotte, D. | | 登録日 | 2013-01-03 | | 公開日 | 2013-05-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Small molecules inhibit the interaction of Nrf2 and the Keap1 Kelch domain through a non-covalent mechanism.
Bioorg.Med.Chem., 21, 2013
|
|
4IQK
 
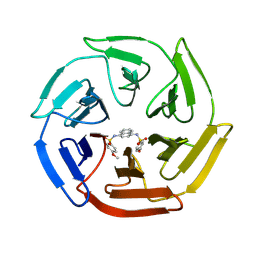 | | Crystal structure of cpd 16 bound to Keap1 Kelch domain | | 分子名称: | Kelch-like ECH-associated protein 1, N,N'-naphthalene-1,4-diylbis(4-methoxybenzenesulfonamide) | | 著者 | Silvian, L, Marcotte, D. | | 登録日 | 2013-01-11 | | 公開日 | 2013-05-15 | | 最終更新日 | 2013-07-03 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Small molecules inhibit the interaction of Nrf2 and the Keap1 Kelch domain through a non-covalent mechanism.
Bioorg.Med.Chem., 21, 2013
|
|
4GWG
 
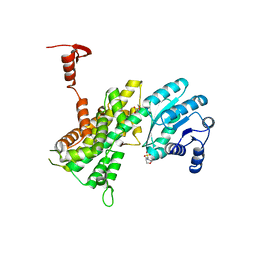 | |
4GWK
 
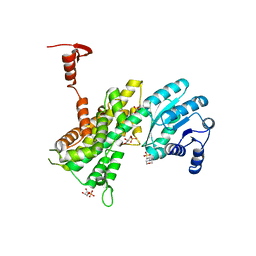 | | Crystal structure of 6-phosphogluconate dehydrogenase complexed with 3-phosphoglyceric acid | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-PHOSPHOGLYCERIC ACID, 6-phosphogluconate dehydrogenase, ... | | 著者 | He, C, Zhou, L, Zhang, L. | | 登録日 | 2012-09-03 | | 公開日 | 2012-11-28 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.534 Å) | | 主引用文献 | Phosphoglycerate mutase 1 coordinates glycolysis and biosynthesis to promote tumor growth.
Cancer Cell, 22, 2012
|
|
5FGP
 
 | | Crystal structure of D. melanogaster Pur-alpha repeat I-II in complex with DNA. | | 分子名称: | CG1507-PB, isoform B, CHLORIDE ION, ... | | 著者 | Weber, J, Janowski, R, Niessing, D. | | 登録日 | 2015-12-21 | | 公開日 | 2016-01-20 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis of nucleic-acid recognition and double-strand unwinding by the essential neuronal protein Pur-alpha.
Elife, 5, 2016
|
|
5FGO
 
 | |
